Table Of ContentEcketal.BMCBioinformatics (2017) 18:441
DOI10.1186/s12859-017-1843-1
RESEARCH ARTICLE Open Access
Interpretation of microbiota-based
diagnostics by explaining individual
classifier decisions
A. Eck1*, L. M. Zintgraf2, E. F. J. de Groot3, T. G. J. de Meij4, T. S. Cohen2, P. H. M. Savelkoul1,6, M. Welling2,5
and A. E. Budding1
Abstract
Background: Thehumanmicrobiotaisassociatedwithvariousdiseasestatesandholdsagreatpromisefornon-invasive
diagnostics.However,microbiotadataischallengingfortraditionaldiagnosticapproaches:Itishigh-dimensional,sparse
andcomprisesofhighinter-personalvariation.Stateoftheartmachinelearningtoolsarethereforeneededtoachieve
thisgoal.Whilethesetoolshavetheabilitytolearnfromcomplexdataandinterpretpatternsthereinthatcannotbe
identifiedbyhumans,theyoftenoperateasblackboxes,offeringnoinsightintotheirdecision-makingprocess.Inmost
cases,itisdifficulttorepresentthelearningofaclassifierinacomprehensibleway,whichmakesthempronetobe
mistrusted,orevenmisused,inaclinicalenvironment.Inthisstudy,weaimtoelucidatemicrobiota-basedclassifier
decisionsinabiologicallymeaningfulcontexttoallowtheirinterpretation.
Results:Weappliedamethodforexplanationofclassifierdecisionsontwomicrobiotadatasetsofincreasingcomplexity:
gutversusskinmicrobiotasamples,andinflammatoryboweldiseaseversushealthygutmicrobiotasamples.The
algorithm simulates bacterial species as being unknown to a pre-trained classifier, and measures its effect on
the outcome. Consequently, each patient is assigned a unique quantitative estimation of which species in their
microbiota defined the classification of their sample. The algorithm was able to explain the classifier decisions
well, demonstrated by our validation method, and the explanations were biologically consistent with recent
microbiota findings.
Conclusions: Application of a method for explaining individual classifier decisions for complex microbiota
analysis proved feasible and opens perspectives on personalized therapy. Providing an explanation to support a
microbiota-based diagnosis could guide decisions of clinical microbiologists, and has the potential to increase
their confidence in the outcome of such decision support systems. This may facilitate the development of new
diagnostic applications.
Keywords: Microbiota, Inflammatory bowel disease (IBD), Supervised classification, IS-pro, Machine learning
Background of microbiota analysis has been extensively studied, for
The human microbiota refers to the trillions of microor- example, for disorders like inflammatory bowel disease
ganismslivingonandwithinourbody,whichareinvolved (IBD) and diverticulitis [3–6] and has led to increasing
inmanybiologicalprocessesnecessarytomaintainhuman effortstodevelopclinicalapplications.
health.Thesemicrobialcommunitiesareassociatedwitha However, using microbiota for diagnostic purposes
widerangeofdiseasestates[1,2],andholdagreatprom- imposes several challenges. Studying the human micro-
ise for non-invasive diagnostics. The diagnostic potential biota is an active field of research. As knowledge is still
accumulating, a full biological understanding of the
underlying mechanisms is missing, limiting the diagnos-
*Correspondence:a.eckhauer@vumc.nl tic efforts to data-driven approaches, such as data
1DepartmentofMedicalMicrobiologyandInfectionControl,VUUniversity
mining. The data itself is high-dimensional and has high
medicalcenter,Amsterdam,TheNetherlands
Fulllistofauthorinformationisavailableattheendofthearticle
©TheAuthor(s).2017OpenAccessThisarticleisdistributedunderthetermsoftheCreativeCommonsAttribution4.0
InternationalLicense(http://creativecommons.org/licenses/by/4.0/),whichpermitsunrestricteduse,distribution,and
reproductioninanymedium,providedyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinkto
theCreativeCommonslicense,andindicateifchangesweremade.TheCreativeCommonsPublicDomainDedicationwaiver
(http://creativecommons.org/publicdomain/zero/1.0/)appliestothedatamadeavailableinthisarticle,unlessotherwisestated.
Ecketal.BMCBioinformatics (2017) 18:441 Page2of13
inter-personal variability. Consequently, identification of the diagnostic Porto-criteria for pediatric IBD. Fresh
meaningful patterns isthwarted. fecal samples were collected in sterile containers and
Therefore, machine learning (ML) tools are particu- immediately stored in the freezer at −20 °C as was
larly useful in this field. Compared to humans, classifiers previouslydescribed[9].
can more easily identify patterns in high-dimensional
data, being able to learn to distinguish between categor- MicrobiotaprofilingbyIS-pro
ies (e.g., sick vs. healthy) based on examples. A trained All samples were analyzed by intergenic spacer (IS)
classifier can then make predictions for new patients, profiling (IS-pro) [8, 9]. IS-pro is a PCR-based technique
accompanied by a probability reflecting its certainty. that differentiates bacterial species by the length of the
However, contrary to humans, classifiers usually do not 16S–23S rRNA in combination with phylum-specific
provide an explanation to their propositions. In most fluorescently labeled PCR primers. Three phylum-
cases they are considered as black boxes, taking features specific primers were used: (1) Bacteroidetes, (2) Firmi-
(e.g., microbial species) as input and returning a predic- cutes, Actinobacteria,Fusobacteria,andVerrucomicrobia
tion. In a diagnostic setting, ML tools are able to assist (FAFV), and (3) Protebacteria. For more details see the
us,but still cannotbeblindly trusted. Additionalfile1:SupplementaryMethods.
Therefore, in order to effectively incorporate ML tools
in microbiota-based diagnostics it is important to pro- Preprocessing
vide clinical microbiologists with a sensible, sample- Preprocessing was carried out with the IS-Pro propri-
specific explanation for every classifier prediction. This etary software suite (IS-Diagnostics, Amsterdam, the
will allow them to weigh the outcome in the clinical and Netherlands) and resulted in microbial profiles, pre-
biological context, interpret it and even adjust specific sented as peak profiles. Each peak represents an IS frag-
therapy based on microbial species that appear to play a ment and is characterized by a color that corresponds to
role inthediagnosis. the phylum (or phyla). The length of the IS fragment,
In this paper, we present how an idea originally pro- measured in nucleotides (nt), discriminates bacterial
posed by Robnik-Šikonja and Kononenko [7] can be species, and its intensity, measured in relative fluores-
implemented for microbiota-based diagnostics. The cence units (RFU), reflects the abundance. Intensity
method simulates input features as being unknown to values were log2 transformed. Each peak was taken as a
measure their effect on the classification outcome. Con- feature for classification. After excluding all-zero fea-
sequently, each input feature is assigned a relevance tures, 914 and 1199 features were left for classification
value that reflects its importance for a specific decision intheSVGandIBDdatasets,respectively.
of the classifier. With this approach, we aim to provide
each patient a unique quantitative estimation of which Supervisedclassification
species in their microbiota were used to determine their The following classifiers were used: Linear Support
health status by the classifier. Given a trained classifier Vector Machine (SVM), Random Forest (RF), Nearest
and a microbiota sample of a new patient, the species Shrunken Centroids (NSC) and Logistic Regression with
relevance ranking may help interpret why this patient L2regularization (LR).
was given a specific diagnosis, highlight species of inter- These classifiers are especially suitable for high-
est,andguidetreatment. dimensionaldataastheyapplyregularizationordimension-
This method was applied on two microbiota datasets ality reduction. Examples of their applicability to human
generated by IS-pro, a technique designed for micro- microbiotawerepreviouslydescribed[10].Implementation
biota profiling in clinical routine [8]. We further also was done using packages of the scikit-learn 0.17.0 Python
describe how the calculated explanations can be vali- library: svm.SVC (with linear kernel and probabilistic
dated and visualized. outputs); neighbors.nearest_centroid.NearestCentroid (with
a 0.1 shrink threshold and probabilities as computed in
Methods [11]);ensemble.RandomForestClassifier(with50trees),and
Subjectsandsamples linear_model.LogisticRegression(withL2penalty).
Two balanced datasets were included: skin versus gut Overall relevance measures of NSC were obtained per
microbiota (SVG, n = 94) and gut microbiota of IBD class by taking the difference between the global
patients versus healthy individuals (IBD, n = 112). Skin centroid and the class centroid, to which we compared
microbiota samples were collected by swabbing the our explanation for the predicted class. For the RF
inner lower arm of healthy adults. Swabs were stored in classifier, we used the importance ranking given by
RTF buffer immediately after collection and later frozen RandomForestClassifier.feature_importances. Note that
at −20 °C. Gut microbiota was sampled from healthy using the weights of a linear SVM as an indication of
children and children diagnosed with IBD according to feature importance is wrong and misleading [12].
Ecketal.BMCBioinformatics (2017) 18:441 Page3of13
Similarly, the coefficients of the LR classifier cannot be relevance of the respective input feature. A positive
interpreteddirectly. relevance value means that the feature value is a
supportive evidencefor the decision, andanegative rele-
Algorithmforexplainingclassifierdecisions vance value expresses that the feature value constitutes
Robnik-Šikonja and Kononenko [7] proposed that in evidence against the decision (and therefore evidence for
order to measurehow important afeature (e.g., abacter- theotherclassinatwo-classproblemlikeours).
ial species) is, we can evaluate the classifier output when
that feature is considered “unknown”. In our case, we Univariatevs.multivariateapproach
seek to evaluate the decision the classifier would make if The method described above is a univariate approach:
the information about a single bacterial species is not to estimate a feature’s relevance, we marginalized out
available. By simulating this for every feature individu- only this one feature. Intuitively, we would expect the
ally, we obtain an overall ranking of the importance of classifier to stay robust to changes in only one feature,
eachspeciesfor a sample-specificclassification. for example when features are redundant. In this case, it
The authors proposed three strategies to evaluate the might be necessary to remove several features at once to
classifier while leaving one feature out: simply declare have a noticeable effect on the prediction of the classi-
the feature as unknown (which only few classifiers fier. We propose a computationally feasible multivariate
allow), re-train the classifier without that feature (which implementation for microbiota data in the Additional
is computationally unfeasible for clinical applications if file1:SupplementaryMethods.
there are many features), or approximately marginalize
the feature out. We chose the latter for our analysis. Analyticvalidationofresultingexplanations
Based on this strategy, we used the empirical distribu- When thedata is poorly understood byhumans,it isnot
tion of the feature in the dataset, and replaced its value straightforward to assess how good an explanation is.
in the given sample with all other possible values it We propose a novel method for the analytic validation
obtained in the dataset. The average prediction probabil- of the explanations, since we have not found any such
ity over those values was then used to determine the method in literature. The relevance estimation method
importance of the feature (see the Additional file 1: described above returns a relevance ranking which indi-
Supplementary Methods for the mathematical formula- cates a contribution measure of each feature. Features
tion). The algorithm returns a vector of the same size as were sorted according to their relevance in ascending
the number of features, where each entry reflects the order (from features with negative influence on the class
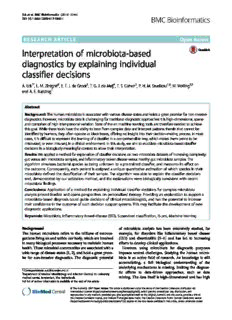
A
B C
D E
Fig.1Agutmicrobiotasampleanditspeaks’relevancevalues:(a)Agutmicrobiotasample:Peakscorrespondtobacterialspecies;Peakheights
correspondtoabundance.(b)Relevancevaluescalculatedforeachpresentpeakintheprofilein(a).(c)Relevancevaluescalculatedforeach
zero-valuedpeakintheprofilein(a).(d)Averagegutmicrobiotaprofileacrossallsamples.(e)Averageskinmicrobiotaprofileacrossallsamples;
FAFV:Firmicutes,Actinobacteria,FusobacteriaandVerrucomicrobia;RelevancewascalculatedbasedonSVMpredictions
Ecketal.BMCBioinformatics (2017) 18:441 Page4of13
score, to irrelevant features, to the most important Skinvs.gutmicrobiota
ones). Then, we successively marginalized out a growing This section presents the results for a simple case study:
number of features, starting with the least important theSVGdataset.
ones, until no features were left. We used the predicted
class for the analysis, and observed how the class prob- Explainingasingleprediction
ability changed based on the features' subset used for Figure1displaystheoutputoftheexplanationalgorithm
the classification. The class probability should rise when for a single prediction of the classifier. For this example,
features with negative evidence for the predicted class we selected a gut microbiota sample correctly classified
are ignored, remain stable if features of zero importance by a linear SVM. Bacterial species abundances (feature
are removed, and decline when highly discriminative values) are shown in Fig. 1a, and the relevance values
featuresareignored. calculated for each feature are illustrated below (Fig. 1b
We also assessed our results based on pairwise corre- and c). To facilitate interpretation, we split the relevance
lations between relevance vectors of a sample calculated calculated for positive-valued features (present peaks,
by different classifiers, as well as between sample- Fig. 1b) from that of zero-valued features (absent peaks,
specific relevancemeasures andglobalones. Fig. 1c), since zero-abundance peaks also hold informa-
tion but are not visible in the profile and therefore can
be visually confusing.
Results In general, for a single gut microbiota sample, we
In this study, we calculated explanations for individual expect that common gut colonizers would obtain
classifier decisions using two microbiota datasets. Each positive relevance values when they are found in the
sample is represented as a peak profile, composed of sample, and negative relevance values in case they are
peaks representing different bacterial species and their not detected. On the other hand, common skin colo-
abundances,which aretaken asfeaturesforclassification nizers should obtain negative relevance values when
(see Methods). they are found in the sample and positive values
The first dataset, denoted “SVG” (Skin Versus Gut), otherwise. This was indeed observed for this example,
consisted of 47 skin microbiota samples that were classi- with common members of the gut microbiota, such
fied against 47 gut microbiota samples (both of healthy
subjects). Since themicrobiota composition thatinhabits Table1Mostrelevantbacterialspeciesforclassificationofthe
gutmicrobiotasampleshowninFig.1
skin or gut is very different, this classification task is
relatively easy and can be directly interpreted by experts. Speciesname IS-pro Presence Relevance Common
peak(s) habitat
This dataset was used to demonstrate how the method
Alistipesputredinis 235 + 0.43 Gut
works and how the results can be presented, while
allowing aqualitativeevaluation ofthe method. Alistipesfinegoldii 396 + 0.4 Gut
The second dataset, denoted “IBD”, consisted of 56 231 + 0.33 Gut
IBD gut microbiota samples (20 Ulcerative Colitis Lachnospiraceaesp. 605 + 0.25 Gut
patients and 36 Crohn’s disease patients) classified
Bacteroidesfragilis 537 + 0.22 Gut
against 56 healthy gut microbiota samples. This data is
Staphylococcusepidermidis 313 – 0.2 Skin
more complex and not so well understood, and classi-
Odoribactersplanchnicus 307 + 0.18 Gut
fiers outperform human expertise. It is those cases that
are interesting for clinical applications, and for which ex- Bacteroidessp. 549 + 0.16 Gut
planationsforclassifierdecisionsareespeciallydesirable. Prevotellasp. 438 + 0.15 Gut
The performance obtained by the classifiers in terms Lachnospiraceaesp. 491 + 0.15 Gut
of prediction accuracy in a 10-fold cross-validation test
Alistipesfinegoldii 230 – −0.14 Gut
was: SVM - 98% and 81%, RF - 99% and 81%, NSC -
Bacteroidesvulgatus 478 + 0.14 Gut
99% and 79%, and LR- 100% and 78%, for SVG and IBD
Streptococcusmitis 296 – 0.13 Skin
datasets,respectively.
Each classifier’sdecision isaccompanied byaprobabil- Sutterellawadsworthensis 661 + 0.12 Gut
ity value reflecting the certainty of the classifier (all of unclassifiedBacteroidete 455 + 0.09 Gut
the above classifiers are either probabilistic or their out- unclassifiedProteobacterium 932 + 0.09 –
put can be transformed to probabilities). To explain the
Escherichiacoli 735/828 + 0.08 Gut
decision, we assigned each input feature a relevance
value. A positive value means that the feature’s value was unclassifiedFirmicute 558 + 0.07 Gut
’+’and’-’indicatewhetherapeakispresentorabsentinasample,andneeds
supportiveevidenceforthepredictedclass,andanegative
tobecoupledwiththerelevancevalueforinterpretation.Rankingisbasedon
valuemeansitwasevidenceagainstthepredictedclass. absoluterelevancevalues;ClassificationwasdonebyalinearSVM
Ecketal.BMCBioinformatics (2017) 18:441 Page5of13
as Bacteroides and Prevotella species, being assigned visual interpretation, we again separated the present
positive relevance when they were found in the sam- peaks (Fig. 2a) from the absent peaks (Fig. 2b). In both
ple. This is indicated by the matching peaks between heat maps, the relevance values clustered the samples
Fig. 1b and d. Prevalent skin species that were not according to their (mostly correctly) predicted class.
detected in this sample, such as Staphylococcus epi- Prevalent colonizers of each habitat stood out with es-
dermidis, also obtained positive relevance. This is in- pecially high or low relevance values, depending on the
dicated by the matching peaks between Fig. 1c and e. class and the presence status. Top ranked species, by
Table 1 lists the top ranked species according to ab- means of average absolute relevance across all samples,
solute relevance, together with their presence status are listed in Table 2.
in this sample and the body habitat they usually
colonize as a validation.
Healthyvs.IBDgutmicrobiota
While body habitats are relatively easy to classify, and
Meta-analysisofallsamples the calculated explanations of the classification are suit-
While the proposed method is mainly aimed to explain able for a qualitative interpretation, real-life clinical
single predictions, to validate it and gain insight into its dilemmas are usually much more complicated and much
behavior across the entire dataset, we calculated expla- less straightforward to interpret. The following use-case,
nations for every sample in the dataset. These were in which we classified gut microbiota samples of IBD
summarized in heat maps (Fig. 2) to visualize the over- patients against gut microbiota of healthy individuals, is
all relevance of features in the dataset. To facilitate a agoodexample.
A B
Fig.2Skinmicrobiotavs.gutmicrobiota:Clusteredheatmapsofpeaks’relevancevaluesacrossallsamples.Columnscorrespondtosamples,
rowstopeaks(orbacterialspecies).Toprelevantspeciesforclassificationareindicated.(a)Heatmapincludingonlytherelevancecalculatedfor
positive-abundancepeaksineachsample;(b)Heatmapincludingonlytherelevancecalculatedforzero-abundancepeaksineachsample.Only
peakswithmeanabsoluterelevanceabove0.01areshownineachheatmap.Colorkeyfrompinktoblueindicatesrelevancefromlowtohigh,
respectively.Foreachsamplethepredictedclass(pred)andthetrueclass(obs)areindicated.Clusteringisbasedoncosinecorrelationmatrix
followedbyunweightedpairgroupmethodwitharithmeticmean(UPGMA).RelevancevalueswerecalculatedbasedonlinearSVMpredictions
Ecketal.BMCBioinformatics (2017) 18:441 Page6of13
Table2Mostrelevantbacterialspeciesforclassificationofskinmicrobiotaversusgutmicrobiotasamples
Speciesname IS-propeak(s) Commonhabitat Rankedtop15by:
Alistipesfinegoldii 230/231/395/396/400/407 Gut all
Alistipesputredinis 235/236 Gut all
Bacteroidesvulgatus 478/479 Gut all
Bacteroidesfragilis 537 Gut all
Clostridiumperfringens 235 Gut all
Streptococcusmitis 296/297 Skin all
Staphylococcusepidermidis 312/313 Skin svm,nsc,lr
Prevotellasp. 437/438 Gut svm,rf,lr
Bacteroidessp. 474 Gut svm,rf,nsc
Lactobacillusparacasei 294 Gut svm,nsc
Staphylococcusepidermidis 303 Skin nsc,lr
Streptococcussp. 321 Gut nsc,lr
Bifidobacteriumadolescentis 495 Gut nsc,lr
Clostridiumsp. 231 Gut rf
Odoribactersplanchnicus 308 Gut rf
Streptococcussp. 318 Gut rf
Lactobacillussp. 478 Gut rf
unclassifiedProteobacterium 941 – rf
Lactococcuslactis 344 Gut nsc
Propionibacteriumacnes 269 Skin nsc
Rankingisbasedonabsolutemeanrelevancevaluesacrossallsamplesperclassifier.Speciesareorderedaccordingtothenumberofclassifiersbywhichthey
weretop-ranked.All-allclassifiers;svm-linearSVM;rf-RandomForest;nsc-NearestShrunkenCentroids;lr-L2-regularizedLogisticRegression
Explainingasingleprediction Meta-analysisofallsamples
Figure 3 illustrates the relevance calculated for three Figure5provides anoverviewof the relevance valuescal-
selected IBD individual samples, all correctly classified culatedperpeakforeverysampleinthedataset.Theclus-
by a linear SVM. Similar to Fig. 1, peak-specific tering of the samples matched the predicted classes,
relevance values are presented in reference to their althoughitfittedlessaccuratelythetrue classofthesam-
abundance profile (Fig. 3a and b), which allows a ples, since more samples were misclassified compared to
straightforward interpretation of which peaks in the the SVG dataset. The color pattern for prevalent species
profile drove the classification towards the IBD class or wascomplementarybetweenthetwoclassesandthepres-
against it. In a separate panel (Fig. 3c) the same is ence (Fig. 5a) or absence (Fig. 5b) status, i.e., a prevalent
displayed for zero-abundance peaks. The most relevant peak that stronglysupported one classalsosupported the
species to explain the classifier’s decision for the top otherclassbyitsabsence,andviceversa.Topexplanatory
example arelistedinTable3. speciesbymeansofaverageabsoluterelevanceforclassifi-
In a clinical context, such relevance ranking can cationacrossallsamplesareshowninTable4.
improve the understanding of a classifier’s output by
clinicians, and potentially support their decisions. Discoveringunderlyingsubgroupswithinaclass
Species that were assigned high relevance values may IBD is a complex and multifactorial disease character-
then guide an intervention according to their abun- ized by high between-patient variability in symptoms
dance. For example, in the top case presented in and response to treatment. This variability is attributed
Fig. 3, determining a relevance cutoff of 0.25 would to various factors, including the intestinal microbiota
result in a short list of species that can be presented composition. We show how the calculated explanations
to clinical microbiologists, which is more informative may be used to reveal underlying subgroups within a
than just presenting the decision (i.e. class probabil- group of patients (Fig. 6). Most of Crohn’s disease
ity) itself (Fig. 4). Based on their expertise, it may be patients in our cohort (32/36) were treated with exclu-
advised that eliminating Escherichia coli and/or sup- sive enteral nutrition (EEN), and were evaluated again
plementing Alistipes spp. would be beneficial for this after a six-week period. These patients could be grouped
particular patient. into two clusters based on the calculated relevance
Ecketal.BMCBioinformatics (2017) 18:441 Page7of13
A
B C
D E
Fig.3ThreeIBDgutmicrobiotasamplesandtheircorrespondingpeaks’relevancevaluesillustrateuniquesample-specificrelevanceranking.All
sampleswerecorrectlyclassified,withtheclassifier’sprobabilityvaluesdisplayedpersample.(a)Gutmicrobiotasamples:peakscorrespondto
bacterialspecies;Peakheightscorrespondtoabundance.Classificationprobabilityisshowntotherightofeachprofile.(b)Relevancevalues
calculatedforeachpresentpeakintheprofilesin(a).(c)Relevancevaluescalculatedforeachabsentpeak(i.e.zero-valuedpeak)intheprofiles
in(a).(d)AverageIBDgutmicrobiotaprofileacrossallsamples.(e)AveragehealthymicrobiotaprofileacrossallsamplesFAFV:Firmicutes,
Actinobacteria,FusobacteriaandVerrucomicrobia;RelevancewascalculatedbasedonSVMpredictions
values, with a significant enrichment of patients with thepredictedclass,Fig.7a).Mostfeatureshadanear-zero
induced remission in response to EEN in one cluster relevance score, and were therefore not important for the
comparedtotheothercluster(p=0.04,chi-squaretest). decision. We observed a rise in certainty when features
withhighnegativerelevance wereremoved,followed by a
Analyticvalidationofexplanations strong decline in response to removal of features with
Since the IBD dataset is poorly understood by humans, top-ranked positive evidence (Fig. 7b). In the middle
we sought for a way to analytically validate the results. range, where features with near-zero relevance were ex-
Here, we propose a novel validation method that cluded, the validation curve stagnates, indicating that re-
consists of subsequently removing a growing number of movingthemhadnoinfluenceontheclassification.
features, sorted by their relevance in ascending order. In addition, we validated our results by comparing
For a good explanation, the confidence of the classifier explanations calculated by different classifiers by sam-
is expected to first go up (evidence against the predic- ple, and comparing sample-specific explanations of a
tion is removed), and then start to go down when classifier with the respective model’s global ranking (if
important features are taken out. When all features are available by the classifier). The correlation was higher
removed, the classifier has no information left and there- when calculated within-classifier, i.e. between the
foreitscertaintyshoulddropto0.5inabalanceddataset. sample-specific relevance vectors and the global im-
The average validation results for the IBD dataset are portance ranking of the same classifier, than between
presented in Fig. 7. Only a small number of features was sample-specific relevance vectors of different classi-
very relevant (either as positive or negative evidence for fiers (Additional file 2: Figure S1).
Ecketal.BMCBioinformatics (2017) 18:441 Page8of13
Table3MostrelevantbacterialspeciesforthetopIBD context. Our algorithm is based on a method originally
microbiotasamplepresentedinFig.3 proposedbyRobnik-ŠikonjaandKononenko[7].It gener-
Speciesname IS-propeak(s) Presence Relevance ates a sample-specific feature relevance ranking that,
Escherichiacoli 735 + 0.62 combined with an abundance profile, allows us to
Alistipesfinegoldii 396 – 0.57 reason about the importance of different species in
driving a prediction. This gives an insight into the
Alistipesfinegoldii 230 – 0.52
specific pathogenic perturbations of the microbiota of
unclassifiedProteobacterium 747 + 0.35
individual patients, which in turn may allow adjusting
Lachnospiraceaesp. 606 + 0.3 personalized treatment, supplementing or targeting
Alistipesputredinis 235 – 0.29 specific species.
unclassifiedFirmicute 326 + −0.26 We first tested our approach on an easily classifiable
dataset consisting of gut and skin microbiota samples.
Finegoldiamagna 306 + 0.24
Species ranked highly relevant were identified as
Parabacteroidesdistasonis 461 + 0.23
common inhabitants of each of these body sites [13].
Morganellamorganii 855 + −0.23
Common gut colonizers, such as Bacteroidetes spp. and
Alistipesfinegoldii 400 – 0.21 E. coli, were supportive evidence for the ‘gut’class. Skin
Bacteroidesfragilis 605 + −0.21 samples presented an opposite image, with dominant
Escherichiacoli 827 + 0.19 skin bacteria, such as S. epidermidis and Streptococcus
Alistipesputredinis 236 – 0.18 mitis, driving the classification by their presence. It is
important to note that evidence for or against a class
Streptococcussp. 321 + 0.17
can be found in the presence of certain species, but also
unclassifiedFirmicute 525 + 0.17
intheirabsence.
Bacteroidesvulgatus 479 – 0.16 Assessing the explanations for the ‘IBD versus
Bacteroidessp. 594 + 0.16 healthy’ case was more challenging. Although findings
Alistipesfinegoldii 407 – 0.15 of aberrations in the microbiota composition of IBD
Escherichiacoli 828 – −0.14 patients are well established [14–16], they are not
consistent regarding specific species. Increased
Bacteroidesfragilis 537 – 0.05
amounts of mucosa-associated E. coli, which was
’+’and’-’indicatewhetherapeakispresentorabsentinasample,andneeds
assigned high relevance values by all classifiers tested,
tobecoupledwiththerelevanceforinterpretation.Rankingisbasedon
absoluterelevancevalues;ClassificationwasdonebyalinearSVM were indeed previously detected in IBD patients [17].
We also compared our results to a recent study1 [9]
Discussion in which a core microbiota, prevalent and stable over
In this study, we present how a method aimed to explain time, was identified in healthy children. Interestingly,
individual classifier decisions can be applied to human strong evidence for the IBD class was given by the
microbiota data to facilitate the interpretation of absence of peaks originating from species described
microbiota-based predictions in a biologically meaningful as members of the pediatric healthy core microbiota,
A B
Fig.4Outputofaclassifierwithout(a)andwith(b)explanation
Ecketal.BMCBioinformatics (2017) 18:441 Page9of13
A B
Fig.5IBDgutmicrobiotavs.healthygutmicrobiota:Clusteredheatmapsofpeaks’relevancevaluesacrossallsamples.Columnscorrespondto
samples,rowstopeaks(orbacterialspecies).Mostrelevantspeciesforclassificationareindicated.(a)Heatmapincludingonlytherelevance
calculatedforpositive-abundancepeaksineachsample;(b)Heatmapincludingonlytherelevancecalculatedforzero-abundancepeaksineach
sample.Onlypeakswithmeanabsoluterelevanceabove0.01areshownineachheatmap.Colorkeyfrompinktoblueindicatesrelevancefrom
lowtohigh,respectively.Foreachsamplethepredictedclass(pred)andthetrueclass(obs)areindicated.Clusteringisbasedoncosinecorrelation
matrixfollowedbyunweightedpairgroupmethodwitharithmeticmean(UPGMA).RelevancevalueswerecalculatedbasedonlinearSVMpredictions
such as certain Alistipes, Bacteroides and Prevotella ignored. Indeed, this was observed for all the classifiers
species. These species were mostly absent from IBD we tested in the IBD dataset. We could rank features
samples, and were assigned consistently high rele- correctly according to their relevance even when a
vance values, which resulted in high dendrogram classifier waslesscertainabout itsdecisions andresulted
distances between the two clusters that appear in the in lower probabilities, like in the case of RF (Fig. 7). In
heat map displaying the absent peaks (Fig. 5b). In our case, the set of important features for one individual
general, in all heat maps, clustering based on the cal- sample was quite small, meaning that a short list of the
culated relevance values separated the samples mostly mostimportantspeciescouldbeeffectivelypresentedtoa
according to their predicted class, which means the physician. Note that although the set of important
relevance values explain the classifier decisions well. features was rather small for an individual classification,
To analytically validate this, we proposed a method this doesnot meanthat the classifierrelies only on those,
that estimates how the classifier’s probability for a class and that they hold all the information.The set ofimport-
changeswhenfeaturesof varyingrelevanceareexcluded. antfeaturesvariedwidelybetweensamples(Figs.2and4),
This approach brings value since it is not always which corroborates the need for a sample-specific explan-
straightforward to assess how good an explanation is, ationmethodformicrobiotadata.
especially when we lack knowledge about the domain. A Still, sample-specific explanations were highly corre-
good explanation means that the certainty of a classifier lated with the global feature rankings at the model level
would decline only when high relevance features are (Additional file 2: Figure S1), indicating that our method
Ecketal.BMCBioinformatics (2017) 18:441 Page10of13
Table4MostrelevantbacterialspeciesforclassificationofIBD
gutmicrobiotaversushealthygutmicrobiotasamples
Speciesname IS-propeak(s) Rankedtop15by
Alistipesfinegoldii 230/395/396/400/407 all
Alistipesputredinis 235/236 all
Odoribactersplanchnicus 307/308 all
unclassifiedFirmicute 396 all
Escherichiacoli 735/821/827/828 all
unclassifiedBacteroidete 251 svm,rf,nsc
Lachnospiraceaesp. 491/605 svm,rf,lr
Bacteroidesvulgatus 479 svm,nsc
Lactobacillussp. 478 svm,lr
Streptococcussp. 318 svm,lr
Bacteroidessp. 474 svm,lr
Streptococcussp. 321 nsc,lr
Prevotellasp. 438 rf,nsc
Bacteroidesfragilis 537 rf,nsc
Clostridiumperfringens 233/235 rf,nsc
Lactobacillussp. 244 rf,nsc
unclassifiedFirmicute 156 rf
Rankingisbasedonabsolutemeanrelevancevaluesacrossallsamplesper
classifier.Speciesareorderedaccordingtothenumberofclassifiersbywhich
theyweretop-ranked.All-allclassifiers;svm-linearSVM;rf-RandomForest;
nsc-NearestShrunkenCentroids;lr-L2-regularizedLogisticRegression
conforms to the general rankings. Some discrepancies
occur when comparing explanations between the differ-
ent classifiers, which represent different algorithm
concepts; SVM and LR are linear classifiers, RF is an en- Fig.6Responsetoexclusiveenteralnutrition(EEN)inCrohn’sdisease
semble of decision trees, and NSC relies on the distance patients:aclusteredheatmapdisplayingpeaks’relevancevalues
between data points to class centroids. While there may acrossCrohn’sdiseasepatientswhoreceivedEENtreatment.Columns
correspondtosamples,rowstopeaks(orbacterialspecies).Foreach
be several solutions to a problem and different classifiers
sample,itisindicatedwhetherthepredictionwascorrect(i.e.the
can make different mistakes, we can assume that the samplewasclassifiedas‘IBD’,darkgrey)ornot(thesamplewas
better the prediction accuracy ofa classifier is,the closer classifiedas‘healthy,lightgrey),andwhetherremissionwasinducedin
the explanation of its decision is to the true difference responsetoEEN(green)ornot(red).Theclusterontheleftisenriched
between the classes. Nonetheless, in practice it is also withpatientswithinducedremissioncomparedtotheothercluster
ontheright(p=0.04,chi-squaretest).Onlypeakswithmeanabsolute
important to pay attention to overfitting, which is a
relevanceabove0.01areshown.Colorkeyfrompinktoblueindicates
common concern in small microbiota datasets. While in
relevancefromlowtohigh,respectively.Clusteringisbasedoncosine
our experiments we did not observe overfitting, it is im- correlationmatrixfollowedbyunweightedpairgroupmethodwith
portant to take into account that in such cases the clas- arithmeticmean(UPGMA).Relevancevalueswerecalculatedbasedon
sifier may fit to random noise in the data, and the linearSVMpredictions
explanations mightnotbesensible.
The calculated relevance can also be used to reveal
underlying subgroups within a patient group. As an location, a biomarker to predict the response or to
example, we focused on response to EEN, an effective adjust an individualized application of EEN in chil-
therapeutic measure to induce remission in active dren is not yet available. Our results suggest a role of
Crohn’s disease pediatric patients. EEN consists of a the microbiota in the response to EEN, and illustrate
complete liquid diet [18] and leads to the induction the potential the explanations hold to unveil patterns
of remission in approximately 85% of patients. In our in the data.
cohort, remission was induced in 18 out of 32 pa- Several other techniques also aim to interpret the
tients. While several factors may influence the re- learning of a classifier. These are mostly focused on
sponse to EEN, including disease duration and feature ranking and feature selection, which are global
Description:FAFV: Firmicutes, Actinobacteria, Fusobacteria and Verrucomicrobia; Relevance was calculated based on SVM predictions. Eck et al Common habitat. Ranked top 15 by: Alistipes finegoldii. 230/231/395/396/400/407. Gut all. Alistipes putredinis. 235/236. Gut all. Bacteroides vulgatus. 478/479. Gut all.