Table Of ContentGwinetal.BMCImmunology2013,14:5
http://www.biomedcentral.com/1471-2172/14/5
RESEARCH ARTICLE Open Access
Differential requirement for Hoxa9 in the
development and differentiation of B, NK,
and DC-lineage cells from Flt3+ multipotential
progenitors
Kimberly Gwin, Joseph J Dolence, Mariya B Shapiro and Kay L Medina*
Abstract
Hoxa9 is a homeodomain transcription factor important for the generation ofFlt3+hiIL-7R- lymphoid
biased-multipotential progenitors, Flt3+IL-7R+ common lymphoid progenitors (CLPs), and Bcell precursors (BCP) in
bonemarrow (BM). In addition to B-cell,Flt3+IL-7R+ CLPs possess NKand DC developmental potentials,although
DCsarise from Flt3+IL-7R-myeloid progenitors as well.Inthis study, we investigated therequirement for Hoxa9,
from Flt3+ or Flt3- progenitor subsets, in thedevelopment ofNK and DC lineage cellsin BM. Flt3+IL-7R+Ly6D- CLPs
and their Flt3+IL-7R+Ly6D+ Blineage-restricted progeny (BLP) were significantly reduced inhoxa9−/−mice.
Interestingly, thereduction inFlt3+IL-7R+ CLPs inhoxa9−/− mice had noimpact onthe generation ofNK precursor
(NKP)subsets,thedifferentiation ofNKP into mature NKcells, or NKhomeostasis. Similarly, percentages and
numbers of common dendritic progenitors(CDP), as well as theirplasmacytoid or conventional dendritic cell
progeny inhoxa9−/−mice were comparable to wildtype. These findings reveal distinct requirements for Hoxa9 or
Hoxa9/Flt3 molecular circuits in regulation of Bversus NKand DC development in BM.
Keywords: Hoxa9, Flt3, IL-7R, B-cell, NK-cell, DC, Development
Background transplantation,impairedresponsestolineagedetermining
Cellfatedecisionsinthehematopoieticsystemarecontin- cytokines, and significant reductions in select LSK + sub-
gent upon the concerted activities of transcription factors sets, myeloid precursor subsets, common lymphoid pro-
that activate and/or repress lineage specific developmental genitors (CLP), thymic progenitors, and B cell precursors
programs and signaling molecules that regulate the tem- (BCP)[5-7].Hoxa9regulatesthehematopoieticprogenitor
poralexpressionandactivationstatusoflineageinstructing andBCPpool,inpart,throughtranscriptionalactivationof
regulatory proteins. Hoxa9 is a homeobox transcription the gene encoding Flt3 [4]. The Flt3 signaling pathway is
factorwithkeyrolesinembryogenesis,hematopoiesis,and criticalforregulationofthehematopoieticprogenitorpool
leukemogenesis[1].Hoxa9ishighlyexpressedinprimitive and is essential at various stages for B, DC, and NK deve-
bloodcellsinmouseandman,anddownregulatedasmul- lopmentand/orhomeostasis[4,8,9].
tipotenthematopoieticprogenitorsundergolineagerestric- Downregulationofc-kit,thereceptorforthehematopoietic
tion and commitment [2-4]. The nonredundant role of cytokine stem cell factor, accompanies lymphoid and DC
Hoxa9inhematopoieticprogenitorbiologyhasbeendeter- lineage restriction from Flt3+hi LMPP [10-12]. Lin-c-kit+lo
minedthroughavarietyofloss-andgain-of-functionstra- Flt3+ IL-7R+CLPs are a heterogeneous population that
tegies. Hoxa9−/− mice are viable, but exhibit multiple exhibit B, NK, T, and DC lineage differentiation poten-
hematopoietic defects including impaired recovery from tials. Gene-targeted deletion of hoxa9 significantly
hematopoieticstress,poorcompetitivereconstitutionupon reduces numbers of Flt3+ IL-7R+CLPs and BCP in BM
[8,13-15]. NKP are also the progeny of Flt3+ IL-7R+
*Correspondence:[email protected] CLPs. Importantly, two independent groups using diffe-
DepartmentofImmunology,CollegeofMedicine,MayoClinic,200First rent experimental approaches recently showed that
StreetSW,Rochester,MN55905,USA
©2013Gwinetal.;licenseeBioMedCentralLtd.ThisisanOpenAccessarticledistributedunderthetermsoftheCreative
CommonsAttributionLicense(http://creativecommons.org/licenses/by/2.0),whichpermitsunrestricteduse,distribution,and
reproductioninanymedium,providedtheoriginalworkisproperlycited.
Gwinetal.BMCImmunology2013,14:5 Page2of9
http://www.biomedcentral.com/1471-2172/14/5
hematopoietic progenitors enriched for NK potential Antibodiesandflowcytometry
were IL-7R+, but Flt3-. In both studies, downregulation Methods for flow cytometry and progenitor isolation
of Flt3 and upregulation of CD122 (IL-15Rα) were se- have been extensively described [4,13]. Flow cytometric
quential steps in NK commitment from Flt3+ IL-7R analysis was performed on the FACS-Canto or LSRII
+CLPs [16,17]. CD122 is a critical component of the cytometers (BD Biosciences, San Jose, CA) and ana-
IL-15R complex and IL-15 signaling is essential for NK lyzed using FlowJo software (Tree Star, Ashland, OR).
development [18]. Lin-CD122+ NKP expressed high All antibodies used in this study were purchased from
levels of Id2, a transcription factor important for NK eBioscience, BioLegend, or Pharmingen. Antibody con-
differentiation [16]. Flt3 signaling has been suggested jugations and combinations used to delineate progeni-
to complement IL-15 in regulation of the BM NK cell tor subsets were the following: ALP/BLP stain Lin+
pool as well as NK homeostasis in the spleen [19]. At cocktail (FITC conjugated CD45R/B220, CDllb/Mac1,
present, there is no information regarding the conse- Ly6G/Gr1, TER119, CD3ε, CD8α, CD11c, NK1.1, Ly6C),
quence of Hoxa9-deficiency, through Flt3-dependent or c-kit APCefluor 780, Sca1-PerCP Cy5.5, biotin IL-7R
Flt3-independent regulatory circuits, on NK commit- (visualized with streptavidin PeCy7), Flt3 PE, and Ly6D
ment, differentiation, or homeostasis. APC; NKP stain Lin+ cocktail (APC conjugated CDllb/
Dendriticcells(DC)areessentialinnateimmuneeffector Mac1, CD3ε, NK1.1, CD19, Ly6D), biotin CD244 (visua-
cells.DCsaregeneratedinBMfromboththemyeloidand lized with streptavidin PeCy7), CD27 APCefluor 780, IL-
lymphoid progenitor pathways. Common dendritic pro- 7R PE, Flt3 PerCPefluor 710, and CD122 FITC; and CDP
genitors (CDPs) originate from Flt3+ myeloid progenitors stainLin+cocktail(FITCconjugatedCD45R/B220,CDllb/
that express Flt3+ but lack expression of the IL-7R [12]. Mac1, Ly6G/Gr1, TER-119, CD3ε, CD8α, NK1.1), c-kit
IL-7R+Flt3+ CLP primarily give rise to plasmacytoid DC APCefluor780,Flt3-PE,bio-IL-7R(visualizedwithstrepta-
[20].Flt3signalingisessentialinregulationofDCdevelop- vidin PeCy7), and CD115-APC; NK subsets, CD3ε PeCy7,
ment in BM as well as DC homeostasis in the peripheral CD122 FITC, NK1.1 AF780, and Dx5 APC; DC subsets,
lymphoid organs [9,12,21]. The Ets-family transcription B220 AF780, MHCII APC, and CD11c PerCPCy5.5. All
factor PU.1 regulates Flt3 in DCs [22]. Interestingly, a re- flow cytometric analysis depicts progenitor subsets within
cent study characterizing Hoxa9 target genes found many thelymphocytelightscattergate(LLS)ofBM(Figure1A).
Hoxa9 binding sites were also bound by PU.1, notably flt3
[23]. Therefore, a molecular contribution of Hoxa9 to the Statistics
developmentand/orhomeostasisofDClineagecells,mer- Statistical significance was determined using the Student-t
itsfurtherinvestigation. testandp-valueslessthan0.05wereconsideredsignificant.
ThegoalofthisstudywastoexaminetheroleofHoxa9
in regulation of NK and DC development and differenti- Results
ation. We found that, in contrast to B cells, Hoxa9 is SelectivereductioninFlt3+IL-7R+,butnotFlt3-IL-7R+CLPs
largely dispensable for the development or peripheral inhoxa9−/−mice
homeostasisofNKorDClineagecells.Thesefindingsun- WepreviouslyshowedthatLin-c-kitloIL-7R+Flt3+CLPsare
cover a distinct requirement for Hoxa9 in regulation of B significantly reduced in hoxa9−/− mice [4]. Lin-c-kitloFlt3+
versusNKandDCdevelopmentfromFlt3+MPPs. IL-7R+cells can be fractionated into Flt3+IL-7R+Ly6D- all
lymphoid progenitors (ALP) and Flt3+IL-7R+Ly6D+B line-
Methods age restricted progenitors (BLP) [24]. Therefore, we em-
Mice ployed this flow cytometric analysis to further define the
C57Bl6 mice (8–16 weeks of age) were purchased from lymphoid/B lineage developmental block inhoxa9−/− mice
The Jackson Laboratory (Bar Harbor, ME). Hoxa9−/− [4,15].TheLin+cocktailusedwasextendedtoincludeanti-
and flt3l−/− mice havebeen described [4]and were bred bodies to CD11c, NK1.1, and Ly6C to eliminate contamin-
and maintained at the Mayo Clinic animal facility and ation of CLPs with mature NK1.1+ NK or CD11c/Ly6C+
used between 8–16 weeks of age. All mice were aged DCs. First, Lin-IL-7R+ cells within the lymphocyte light
matched in individual experiments and the experimental scattergate(Figure1A) weregatedforlowexpressionofc-
data did not vary as a function of age or sex. Bone mar- kit (Figure 1B, gated region, middle panel). We note that
row cells from individual male or female mice were col- usingtheextendedLin+cocktailandgatingonIL-7R+cells,
lected from the four hind limb bones after carbon that overall percentages of Lin-IL-7R+ were not reduced in
dioxide asphyxiation and single cell suspensions made in hoxa9−/− mice (0.71±0.20% vs. 0.83±0.29% in hoxa9−/−
preparation for flow cytometric analysis. All experiments vs.wildtype,respectively,n=5mice).TheLin-IL-7R+c-kitlo
were carriedoutinaccordance withMayo ClinicInstitu- subsetwasthenfractionatedintoALPandBLPusingdiffe-
tionalAnimalCareandUseCommitteeguidelinesunder rential expression of Flt3 and Ly6D (Figure 1B, bottom
ProtocolA17509. panels). This gating strategy revealed that ALP, as well as
Gwinetal.BMCImmunology2013,14:5 Page3of9
http://www.biomedcentral.com/1471-2172/14/5
A B C
B6 hoxa9-/-
0.2
s
et
s 0.15
b
R u
7 s
L- P 0.1
I L
C
0.05 * *
%
0
Lin+ ALP BLP Flt3-
Ly6D-
R
7
-
L
I
c-kit
ALP BLP ALP BLP
3
Flt
Flt3-Ly6D- Flt3-Ly6D-
Ly6D
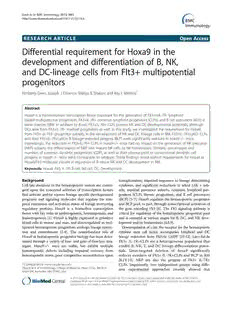
Figure1Reductioninlymphoidprogenitorsubsetsinhoxa9−/−mice.A)Lymphoidlightscattergate(LLS)usedthroughoutthesestudies.
B)FlowcytometricanalysisofBMLin-IL-7R+c-kitloCLPsubsetsinwildtypeB6andhoxa9−/−mice.Thegatingstrategyisdepictedfromtopto
bottomwitharrows.ThelineagecocktailincludesantibodiestoCD45R/B220,CDllb/Mac1,Ly6G/Gr1,TER119,CD3ε,CD8α,CD11c,NK1.1,and
Ly6C.Lin-IL-7R+cells(toppanel)weregatedforIL-7R+c-kitlocells(middlepanel)andLin-IL-7R+c-kitlocellswerefractionatedinto3subsets
basedondifferentialexpressionofFlt3andLy6D.ALPsaregatedasLin-IL-7R+c-kitloFlt3+Ly6D-andBLPasLin-IL-7R+c-kitloFlt3+Ly6D+.Athird
Lin-IL-7R+c-kitloFlt3-Ly6D-subsetislabeledasFlt3-Ly6D-.C)SummaryofALP,BLP,andFlt3-Ly6D-CLPsprecursorfrequenciesinB6(whitebars)
andhoxa9−/−(graybars)mice.Precursorfrequencieswerecalculatedbymultiplyingsequentialpercentagesofeachgatedregion(ie.,%Lin-IL-
7R+inLLSgatex%ckitlox%’sofALP,BLP,orFlt3-Ly6D-).Dataisrepresentativeof5individualB6andhoxa9−/−animalBManalyses.The
asteriskindicatesp<0.05.ErrorbarsrepresentSEM.
their B lineage restricted progeny BLP, are significantly 0.11±0.06% of BM cells within the lymphoid light scatter
reduced in hoxa9−/− mice, consistent with our previous gateinB6vs.hoxa9−/−mice,respectively,p=0.11).Percen-
findings[4].PrecursorfrequencyanalysisrevealedthatALP tages of ALP, BLP, and IL-7R+Flt3-Ly6D- CLPs in B6 con-
represent 0.061±0.013% vs. 0.024±0.01% of BM cells trolandhoxa9−/−micearesummarizedinFigure1C(data
within the lymphoid light scatter gate in B6 vs. hoxa9−/− represents mean and SEM). We conclude from these flow
mice, respectively (p=0.0009). Similarly, BLP represent cytometry analyses that Hoxa9-deficiency preferentially
0.026±0.002% vs. 0.01±0.002% of BM cells within the reducesFlt3+IL-7R+,butnotFlt3-IL-7R+CLPsubsets.
lymphoid light scatter gate in B6 vs. hoxa9−/− mice, re-
spectively (p=0.0005). Less BLP resulted in reductions in NKdevelopmentanddifferentiationinhoxa9−/−mice
CD19+IgM-B cellprecursors(11.5±2%vs.20.03±2.0%of CD27+ and CD244+ are cell surface markers expressed
BM cells within the lymphoid light scatter gate, p=0.0009, on IL-7R-Flt3+ multipotential progenitors (MPP) and
inhoxa9−/−vs.B6,respectively),consistentwithourprevi- CLPs, and maintained on mature NK cells [25,26]. A
ous report [4,15]. In contrast, a conspicuous population of small subset of CD27+CD244+IL-7R+ progenitors were
IL-7R+Flt3-Ly6D- CLPs was not reduced (0.087±0.22% vs. identified in BM that lacked expression of surface
Gwinetal.BMCImmunology2013,14:5 Page4of9
http://www.biomedcentral.com/1471-2172/14/5
markers displayed by lineage committed precursor sub- ofNKP,BMNKtransitionalsubsetsweredistinguishedby
sets, including CD19, Mac1, Ly6D, NK1.1, and CD3. differential expression of CD3ε, CD122, NK1.1, and
CD122 (IL-2Rβ) is a critical component of the IL-15R CD49b/Dx5 [28]. To determine whether Hoxa9 regulates
complex (comprised of IL-15Rα, IL-2Rγ and IL-2Rβ) NK differentiation from NKP, BM cells from B6 wildtype,
[27]. Expression of the IL-15R is acquired as CLP diffe- hoxa9−/−, and flt3−/− mice were stained with antibodies
rentiate into NK cells and is associated with loss of B,T, toCD3ε,CD122,NK1.1,andCD49b(Dx5)[28].Asshown
and DC lineage differentiation potentials. Fathman, in Figure 2D-E, percentages and numbers of CD3ε-CD
et al., evaluated NK precursor potential in Lin- (CD19- 122+NK1.1-Dx5- NKP and CD3ε-CD122+NK1.1+Dx5-
Mac1- Ly6D- NK1.1- CD3-) CD27+CD244+IL-7R+ cells immatureNKcells weresimilar betweenthe 3 genotypes.
[17]. Within the Lin-CD27+CD244+IL-7R+ subset, dif- These results suggest that Hoxa9 is dispensable for NK
ferential expression of Flt3 and CD122 distinguished differentiation from rNKP. However, in contrast to Flt3
three progenitor subsets, Flt3+CD122-, Flt3-CD122-, signaling, Hoxa9 is also dispensable for NK homeostasis
and Flt3-CD122+. Transplantation studies revealed that in BM and spleen as neither mature NK cell frequencies
theLin-CD27+CD244+IL-7R+Flt3+subsetwerefoundto nor numbers are reduced in BM or spleen in hoxa9−/−
functionallylargelyoverlapCLPs,whiletheLin-CD27+and mice(Figure2D-G).
Lin-CD244+subsetsgaverisetoNKcells.Invitrodifferen-
tiation studies confirmed that the Lin-CD27+CD244+ DCdifferentiationinhoxa9−/−mice
IL-7R+Flt3-CD122- and CD122+ subsets generated only Lineage-restricted precursors of DCs are enriched within
NK cells. Based on differential expression of CD122 and Lin-ckit+loFlt3+ MPPs [12]. Flt3 signaling is essential for
the NK lineage bias exhibited by these cells in functional DC differentiation and homeostasis [21,29]. Lineage-
assays, the Lin-CD27+CD244+IL-7R+Flt3-CD122- subset restricted common dendritic progenitors CDP express
wasdesignatedPreNKPandtheLin-CD27+CD244+IL-7R+ theCD115/M-CSFR,butlackexpressionofthelymphoid-
Flt3-CD122+ subset rNKP [17]. Previous studies showed lineage-associated cytokine receptor IL-7R [12]. Lin-
that loss of Flt3-L significantly reduced the mature BM ckit+loFlt3+ progenitors are reduced in hoxa9−/− mice
NK pool [9,19]. However, a role for Hoxa9, through Flt3 (Figure 3A, middle panel). However, percentages of
dependent or independent mechanisms, in regulation of Lin-c-kit+lo Flt3+ cells expressing the CD115/M-CSFR
NK commitment and differentiation from the CLP stage were comparable between wildtype and hoxa9−/− mice
hasnotbeendetermined. (Figure 3A and summarized in Figure 3B). Next we com-
WeshowedinFigure1B-CthatasubsetofIL-7R+CLPs pared percentages and absolute numbers of DC-lineage
thatlackFlt3-andLy6D-aresparedbyHoxa9-deficiency. progeny. CD11c+MHCII+B220- conventional DC (cDC)
Since IL-7R+Flt3-Ly6D- CLPs are enriched for NK pre- and CD11c+MHCII+B220+ plasmacytoid DC (pDC) are
cursors we speculated that Hoxa9 might be dispensable the progeny of CDP, although there is experimental evi-
for NK commitment and differentiation from CLPs. To dence that DCs develop from CLP [20,29-31]. As shown
make this determination, we employed the flow cyto- inFigure4A-C,frequenciesandabsolutenumbersofcDC
metric analysis described above to examine the NK pre- (gray bars) and pDC (white bars) in hoxa9- mice were
cursor compartment in hoxa9−/− mice. Consistent with largely comparable to WT. Furthermore, percentages and
our findingsin Figure1B, percentages and absolute num- absolute numbers of cDC and pDC were also found at
bers of Lin- CD27+CD244+IL-7R+Flt3+ CLPs were sig- comparablefrequenciestoB6wildtypemiceinthespleen
nificantly reduced in hoxa9−/− mice (Figure 2A-C). (data not shown). Consistent with previous findings, cDC
Importantly, shown in Figure 2A and summarized in and pDC were reduced in BM and spleen of flt3l−/− ani-
Figures2B-C,neitherfrequenciesnornumbersofPreNKP mals (Figure 4B-C and data not shown) [9]. These data
or rNKP within the lymphoid light scatter gate in BM suggest that Hoxa9 is not a critical component of the
were diminished by Hoxa9-deficiency. The reduction in regulatory circuitry that orchestrates DC commitment
CLPs reinforces the requirement for Hoxa9 in regulation and/ordifferentiationfromFlt3+MPPs.
oftheIL-7R+Flt3+CLPsubset.Surprisingly,thereduction
in IL-7R+Flt3+ CLPs did not impact the generation or Discussion
maintenance of Pre-NKP or rNKP. These data suggest In this study we investigated the role of Hoxa9 in regula-
that commitment to the NK fate is not dependent on tion of NK- and DC-lineage commitment and differenti-
Hoxa9functionwithinCLPs. ation in BM. Hoxa9-deficient mice exhibit reductions in
Flt3 signaling has been implicated in regulation of the select hematopoietic progenitor subsets, including Flt3+
mature BM NK compartment. Specifically, it was shown MPPs, Flt3+IL-7R+ CLPs, myeloid progenitor subsets,
that mature Lin-CD122+NK1.1+Dx5+ NK cells are and BCPs, in addition to Pro-T subsets in the thymus
reduced in flt3l−/− mice and our results concur with [4,15]. Hoxa9 regulates these hematopoietic progenitor
those findings (Figure 2D-E) [19]. Until the identification subsets,atleastinpart,throughtranscriptionalregulation
Gwinetal.BMCImmunology2013,14:5 Page5of9
http://www.biomedcentral.com/1471-2172/14/5
A B D
FSC B6 hoxa9-/- M Precursor frequency 000000...000...000012123555 * Bho6xa9-/- BM Precursor frequency 012...12555 Bhflto63xl-a*/9--/-
B 0 0
CLP PreNKP rNKP
Lin+
4
4 C E
2
CDCD27 S per 4 hindlegbones 112250505000000000000000 * Bho6xa9-/- #’s per 4 hindlegbonesX10-60011....4826 Bhflto63xl-a/9--/*-
#’ 0 CLP PreNKP rNKP 0
C
S F
F B6 hoxa9-/-
C
S
S G
IL7R
25
5
0-
CLP CLP CD3ε # x1 20 ns
Flt3 K1.1 NK cell 1105
N n
ee 5
PreNKP rNKP PreNKP rNKP pl
S 0
B6 hoxa9-/-
CD122 CD122
Figure2Hoxa9isdispensableforNKdifferentiationandhomeostasis.A)BMcellsfromwildtypeB6orhoxa9−/−micewereisolatedand
stainedwithcombinationsofantibodiesasdetailedintheMethodstoresolvePreNKPandrNKPprogenitorsubsets.TheLin+antibodycocktail
includesCDllb/Mac1,CD3ε,NK1.1,CD19,andLy6D.Thegatingstrategyisdepictedfromtoptobottomwitharrows.Dataisrepresentativeof6
individualmiceofeachgenotype.B)NKPsubsetfrequenciesinBMofwildtypeB6(openbars)andhoxa9−/−(graybars)mice.Precursor
frequenciesreflectpercentagesofCLP,PreNKP,orrNKPwithintheLin-CD244+CD27+IL-7R+gatedfractions.C)AbsolutenumbersofBMNKP
subsetsinwildtypeB6(openbars)andhoxa9−/−(graybars)mice.D)SummaryoffrequenciesorE)absolutenumbersofBMtotalCD3ε-CD122+
progenitors,NKP(CD3ε-CD122+NK1.1-Dx5-),immatureNK(CD3ε-CD122+NK1.1+Dx5-),andmatureNK(CD3ε-CD122+NK1.1+Dx5+)cellsin
wildtypeB6(openbars),hoxa9−/−(lightgraybars),andflt3l−/−(darkgraybars).Datareflectstheaverageofeachindicatedsubsetpooledfrom
analysisof7–8micepergenotype.F)FlowcytometryprofileofCD3ε-NK1.1+CD122+NKcellsinspleen.Spleenmononuclearcellswerefirst
gatedonCD3ε-(toppanels).TheCD3ε-splenocyteswerethenanalyzedforexpressionNK1.1andCD122(bottompanels).G)Absolutenumbers
ofNKcells(CD3ε-NK1.1+CD122+)inspleen.Precursorfrequencieswerecalculatedbymultiplyingsequentialpercentagesofeachgatedregionas
describedinFigure1.AbsolutenumbersweredeterminedbymultiplyingprecursorfrequenciesxnumbersofBMmononuclearcellsobtained
from4hindlimblegbones.Theasteriskindicatesp<0.05.ErrorbarsrepresentSEM.
ofFlt3[4].Flt3signalingisimportantforthedevelopment Flt3+IL-7R+ CLPs, as well as their Flt3+IL-7R+Ly6D+
and/or maintenance of Flt3+ MPPs, CLPs, B,T, NK, and B-lineagerestrictedprogeny.Importantly,thereductionin
DC lineage cells [9,19]. Previously, a role for Hoxa9, Flt3+IL-7R+ CLPs in hoxa9−/− mice does not comprom-
through Flt3-dependent or independent regulatory cir- ise the generation of NKP. These findings distinguish a
cuits, in regulation of NK- or DC-lineage commitment criticalfunctionforHoxa9inFlt3+IL-7R+CLPsinregula-
and differentiation in BM had not been established. Here tionofB,butnotNKlineagecommitment.
weshowthatHoxa9isdispensableforthegenerationand The differentiation of DC lineage cells from multipo-
differentiationofNKand DC lineage cellsinBM. Incon- tent hematopoietic progenitors is driven by the com-
trast, Hoxa9 plays a non-redundant role in regulation of binatorial activities of cytokines, notably Flt3L, M-CSF,
Gwinetal.BMCImmunology2013,14:5 Page6of9
http://www.biomedcentral.com/1471-2172/14/5
A B
B6 hoxa9-/-
0.15
P
C CD 0.1 ns
S M
F B
% 0.05
0
Lin+ B6 hoxa9-/-
kit
c-
Flt3
CDP CDP
5
1
1
D
C
IL7R
Figure3Hoxa9isdispensableforthedevelopmentofCDPinBM.A)FlowcytometryprofileofCDPinB6andhoxa9−/−mice.TheLin+
antibodycocktailincludedCD45R/B220,CDllb/Mac1,Ly6G/Gr1,TER-119,CD3ε,CD8α,andNK1.1.Thegatingstrategyisdepictedfromtoptobottom
witharrows.CDPareidentifiedasLin-ckitloFlt3+CD115/M-CSFR+.B)CDPprecursorfrequency.Precursorfrequencieswerecalculatedbymultiplying
sequentialpercentagesofeachgatedregionasdescribedinFigure1.Dataisrepresentativeof4individualmicepergenotype.ErrorbarsrepresentSEM.
and GM-CSF [32]. Importantly, DCs develop from both progenitors are generated in flt3−/− and flt3l−/− mice,
myeloid and lymphoid precursors. Myeloid progenitors albeit at reduced frequencies [8,13]. Thus, Flt3 signaling
are minimally affected by Hoxa9-deficiency, thus provi- is not essential for induction of IL-7R expression, con-
ding an alternate route for DC production [6]. Although sistent with our current findings. CD122 is upregulated
Hoxa9 is a critical regulator of flt3, Flt3 is expressed, al- in Flt3- CLPs uncoupling Flt3 signaling from induction
beit at lower levels, in Hoxa9-deficient hematopoietic of CD122 expression. PreNKP have been proposed to be
progenitors [4]. In addition to Hoxa9, the Ets-family developmentally positioned downstream of Flt3+ IL-7R+
transcription factor PU.1 is a key regulator of flt3. It was CLPs (Figure 5). We note that the few B220+ cells iden-
previously established that PU.1 regulation of Flt3 is es- tified in FL−/− x IL-7Ra−/− mice are NK1.1+ [36]. Fur-
sential for DC development [22]. Hoxa9 transcripts are thermore, il7−/− mice exhibit no deficiencies in NK
low/negative in CDP and Macrophage Dendritic Pro- cells in BM [19,37]. These experimental results together
genitors (MDP), compared to Flt3+ MPP or CLPs. In with ourfindingsherein suggestthatcommitment to the
contrast, PU.1 transcripts are elevated in CDP and MDP NK fate is not dependent on the combinatorial activities
(Figure 5) [33]. Hierarchically, MDP give rise to CDP, ofFlt3and/orIL-7R signalingorHoxa9function.
andCDParethelineage restrictedprecursorsofconven- We show significantly reduced percentages of Flt3+IL-
tional and plasmacytoid dendritic cells [34]. Interes- 7R+ CLP/ALPs and Flt3+IL-7R+BLPs in hoxa9−/− mice.
tingly, MDP numbers are not reduced in flt3−/− mice, The combinatorial activities of Flt3 and IL-7R signaling
suggesting that Flt3 expression is not required for com- areindispensableforBcellgenesis[36].IL-7Rsignalingin
mitment to the DC fate [21]. Our data extend these CLPs promotes the generation of BLP and their subse-
results and establish that Hoxa9 function is dispensable quent differentiation into BCP through Stat5-mediated
forcommitment anddifferentiationintheDClineage. induction of the B-cell fate determinant EBF [38,39]. EBF
A recent fate mapping study of an IL-7R reporter in turn suppresses Id2 transcription [40,41]. Id2 is critical
mouse suggested that all NK-lineage cells progress for NK differentiation and transcript levels are increased
through an IL-7R+stage [35]. IL-7R+hematopoietic in IL-7R+Flt3- rNKP [16,42-44]. The observation that
Gwinetal.BMCImmunology2013,14:5 Page7of9
http://www.biomedcentral.com/1471-2172/14/5
A
Lymphocyte light scatter MHCII+ MHCII + B220- MHCII + B220+
r
e
tt cDC pDC
a
c
s
B6
d
r
a
w
r
o
F
r
e
t
t
a cDC pDC
c
s
d flt3l-
r
a
w
r
o
F
r
e
t
t
a
c cDC pDC
s
d
r hoxa9-/-
a
w
r
o
F
MHCII B220 CD11c CD11c
B C
3 250
s 1.2 -
t 0
e 1
s 1 200
b x
u s
C s 0.8 set 150
M D 00..46 * ns C sub 100 * *
B
D 50
% 0.2
M
B
0 0
B6 hoxa9-/- flt3l-/- # B6 hoxa9-/- flt3l-/-
Figure4Hoxa9isnotrequiredforDCdifferentiation.A)FlowcytometriccomparisonofDCsubsetsinBMofwildtypeB6,flt3l−/−,and
hoxa9−/−mice.BMcellswerefirstanalyzedforMHCIIexpression.TheMHCII+cellswerethenfractionatedbasedondifferentialexpressionof
CD45R/B220intoMHCII+B220+orMHCII+B220-.ConventionalDCsareMHCII+B220-CD11c+.PlasmacytoidDCsareMHCII+B220+CD11c+.B)pDC
(whitebars)andcDC(graybars)precursorfrequencyinBM.C)AbsolutenumbersofpDC(whitebars)andcDC(graybars)inBM.Precursor
frequencieswerecalculatedbymultiplyingsequentialpercentagesofeachgatedregionasdescribedinFigure1.Absolutenumberswere
determinedbymultiplyingprecursorfrequenciesxnumbersofBMmononuclearcellsobtainedfrom4hindlimblegbones.Datais
representativeof4micepergenotype.ErrorbarsrepresentSEM.
Gwinetal.BMCImmunology2013,14:5 Page8of9
http://www.biomedcentral.com/1471-2172/14/5
Hoxa9+ Hoxa9-lo Hoxa9-
PFILLylU-t637.D1+R+++ PFILlU-t37.1-R++ PFILlU-t37.1-R++
Hoxa9+ Hoxa9+ Hoxa9+ Hoxa9+ Hoxa9+ BLP ProB PreB
PU.1+lo PU.1+ PU.1+ PU.1+ PFlUt3.1++hi
Flt3- Flt3+lo Flt3+ Flt3+hi IL-7R+
IL-7R- IL-7R-lo Ly6D-
HSC MPP GMLP LMPP CLP/ALP
Hoxa9- Hoxa9- Hoxa9- Hoxa9-
PU.1+ PU.1+ PU.1+ PU.1+
Flt3-CD122- IL-7R+ Flt3- Flt3-
IL-7R+ CD122+ CD122+ CD122+
Hoxa9+lo
PU.1+ PreNKP rNKP imNK mNK
Flt3+
MDP
PU.1+
Flt3+
M-CSFR+
IL-7R-
CDP
Figure5ModeldepictingHoxa9andFlt3expressionpatternsinDC,NK,andBcellprecursorsinBM.HSC,MPP,GMLPandLMPPare
primitiveLSK+subsetsdefinedbydifferentialexpressionofFlt3andVCAM-1[46].CLPareLin-ckitloIL-7R+Flt3+Ly6D-.BLPsareaBlineage
restrictedsubsetofCLP/ALPthatexpressLy6D.ProBthroughPreBareB220+Bcellprogenitorsubsets.PreNKPareLin-CD244+CD27+IL-7R+Flt3-
loCD122-andarethepresumedprogenyofCLP/ALP.rNKPthroughmNKareNKlineagerestrictedsubsetsdistinguishbasedonexpressionof
CD122,NK1.1andDx5.Hoxa9andFlt3expressionpatternswithinthesubsetsarederivedfromourpreviousstudy[4],ordeducedfromthe
ImmunologicalGenomicProjectdatabase(http://www.immgen.org/databrowser/index.html).
frequenciesandnumbersofPreNKPandrNKPareessen- Abbreviations
tially unchanged while BLP are reduced suggest that BM:Bonemarrow;MPP:Multipotentialprogenitor;LSK:LineagenegativeSca-
1+c-kit+hi;GMLP:Granulocyte-macrophage-lymphoidprogenitor;
Hoxa9,alone or in combination withFlt3 signaling, regu-
LMPP:Lymphoidbiasedmultipotentialprogenitor;CLP:Commonlymphoid
lates a critical molecular circuitry that dictates the B vs. progenitor;ALP:Alllymphoidprogenitor;BCP:Bcellprecursor;NKP:Natural
NKfatedecisioninFlt3+IL-7R+CLP/ALPs. killercellprogenitor;rNKP:Restrictednaturalkillercellprogenitor;
CDP:Commondendriticprogenitor;MDP:Macrophagedendriticprogenitor;
Finally, the dramatic downregulation of Flt3 in PreNKP
Flt3L:Flt3ligand.
suggests that commitment to the NK fate might initiate a
series of events that silences flt3 expression. Induction of Competinginterests
the B lineage commitment factor Pax5 silences Flt3 tran- Theauthorsdeclarethattheyhavenocompetingfinancialinterests.
scription in committed BCP [45]. Interestingly, failure to
Authors’contributions
downregulate Flt3 impairs B cell differentiation. Ectopic
KLMconceivedanddesignedtheexperiments,performeddataanalysis,
expression studies examining the consequences of sus- madefiguresandwrotethemanuscript.KG,JJD,MBSdesignedand
tained expression of Flt3 or Hoxa9 on NK differentiation performedexperimentsandanalyzeddata.Allauthorsreadandapproved
thefinalmanuscript.
will be informative with regard to the requirement for
regulatedexpressionofFlt3orHoxa9inNKdevelopment. Acknowledgements
TheauthorsthankDr.VirginiaSmithShapiroforhelpfulcommentsonthe
manuscript.K.L.M.issupportedbyR01HL096108fromtheNationalHeart,
Conclusion
LungandBloodInstitute.
In summary, Hoxa9 is a critical regulator of early
hematopoietic differentiation, in part through regulation Received:16October2012Accepted:25January2013
Published:30January2013
of Flt3. We uncovered differential requirements for a
Hoxa9 in the development/maintenance of IL-7R+Flt3+
References
CLPs and BLP. Although NKP and CDP share a com- 1. AbramovichC,HumphriesRK:Hoxregulationofnormalandleukemic
mon Lin-c-kitloFlt3+ progenitor with BLP, Hoxa9 is dis- hematopoieticstemcells.CurrOpinHematol2005,12(3):210–216.
2. SauvageauG,LansdorpPM,EavesCJ,HoggeDE,DragowskaWH,ReidDS,
pensable for their development and differentiation.
LargmanC,LawrenceHJ,HumphriesRK:Differentialexpressionof
These findings reinforce the stringent requirement for homeoboxgenesinfunctionallydistinctCD34+subpopulationsofhuman
Hoxa9 in regulation of B lymphopoiesis and support fu- bonemarrowcells.ProcNatlAcadSciUSA1994,91(25):12223–12227.
3. SoCW,KarsunkyH,WongP,WeissmanIL,ClearyML:Leukemic
ture studies aimed at delineating Hoxa9-dependent gene-
transformationofhematopoieticprogenitorsbyMLL-GAS7inthe
ticcircuitsthatdictatetheBcellfatedecision. absenceofHoxa7orHoxa9.Blood2004,103(8):3192–3199.
Gwinetal.BMCImmunology2013,14:5 Page9of9
http://www.biomedcentral.com/1471-2172/14/5
4. GwinK,FrankE,BossouA,MedinaKL:Hoxa9regulatesFlt3in 25. WiesmannA,PhillipsRL,MojicaM,PierceLJ,SearlesAE,SpangrudeGJ,
lymphohematopoieticprogenitors.JImmunol2010,185(11):6572–6583. LemischkaI:ExpressionofCD27onmurinehematopoieticstemand
5. LawrenceHJ,ChristensenJ,FongS,HuYL,WeissmanI,SauvageauG, progenitorcells.Immunity2000,12(2):193–199.
HumphriesRK,LargmanC:LossofexpressionoftheHoxa-9homeobox 26. KimS,IizukaK,KangHS,DokunA,FrenchAR,GrecoS,YokoyamaWM:In
geneimpairstheproliferationandrepopulatingabilityofhematopoietic vivodevelopmentalstagesinmurinenaturalkillercellmaturation.Nat
stemcells.Blood2005,106(12):3988–3994. Immunol2002,3(6):523–528.
6. LawrenceHJ,HelgasonCD,SauvageauG,FongS,IzonDJ,HumphriesRK, 27. BudagianV,BulanovaE,PausR,Bulfone-PausS:IL-15/IL-15receptor
LargmanC:Micebearingatargetedinterruptionofthehomeoboxgene biology:aguidedtourthroughanexpandinguniverse.CytokineGrowth
HOXA9havedefectsinmyeloid,erythroid,andlymphoidhematopoiesis. FactorRev2006,17(4):259–280.
Blood1997,89(6):1922–1930. 28. RosmarakiEE,DouagiI,RothC,ColucciF,CumanoA,DiSantoJP:
7. ThorsteinsdottirU,MamoA,KroonE,JeromeL,BijlJ,LawrenceHJ, IdentificationofcommittedNKcellprogenitorsinadultmurinebone
HumphriesK,SauvageauG:Overexpressionofthemyeloidleukemia- marrow.EurJImmunol2001,31(6):1900–1909.
associatedHoxa9geneinbonemarrowcellsinducesstemcell 29. KarsunkyH,MeradM,CozzioA,WeissmanIL,ManzMG:Flt3ligandregulates
expansion.Blood2002,99(1):121–129. dendriticcelldevelopmentfromFlt3+lymphoidandmyeloid-committed
8. MackarehtschianK,HardinJD,MooreKA,BoastS,GoffSP,LemischkaIR: progenitorstoFlt3+dendriticcellsinvivo.JExpMed2003,198(2):305–313.
Targeteddisruptionoftheflk2/flt3geneleadstodeficienciesin 30. TraverD,AkashiK,ManzM,MeradM,MiyamotoT,EnglemanEG,Weissman
primitivehematopoieticprogenitors.Immunity1995,3(1):147–161. IL:DevelopmentofCD8alpha-positivedendriticcellsfromacommon
9. McKennaHJ,StockingKL,MillerRE,BraselK,DeSmedtT,MaraskovskyE, myeloidprogenitor.Science(NewYork,NY2000,290(5499):2152–2154.
MaliszewskiCR,LynchDH,SmithJ,PulendranB,etal:Micelackingflt3 31. ManzMG,TraverD,MiyamotoT,WeissmanIL,AkashiK:Dendriticcell
ligandhavedeficienthematopoiesisaffectinghematopoieticprogenitor potentialsofearlylymphoidandmyeloidprogenitors.Blood2001,
cells,dendriticcells,andnaturalkillercells.Blood2000,95(11):3489–3497. 97(11):3333–3341.
10. KondoM,WeissmanIL,AkashiK:Identificationofclonogeniccommon 32. BelzGT,NuttSL:Transcriptionalprogrammingofthedendriticcell
lymphoidprogenitorsinmousebonemarrow.Cell1997,91(5):661–672. network.NatRevImmunol2012,12(2):101–113.
11. PayneKJ,MedinaKL,KincadePW:Lossofc-kitaccompaniesB-lineage 33. MillerJC,BrownBD,ShayT,GautierEL,JojicV,CohainA,PandeyG,Leboeuf
commitmentandacquisitionofCD45RbymostmurineB-lymphocyte M,ElpekKG,HelftJ,etal:Decipheringthetranscriptionalnetworkofthe
precursors.Blood1999,94(2):713–723. dendriticcelllineage.NatImmunol2012,13(9):888–899.
12. OnaiN,Obata-OnaiA,SchmidMA,OhtekiT,JarrossayD,ManzMG: 34. LiuK,VictoraGD,SchwickertTA,GuermonprezP,MeredithMM,YaoK,ChuFF,
IdentificationofclonogeniccommonFlt3+M-CSFR+plasmacytoidand RandolphGJ,RudenskyAY,NussenzweigM:Invivoanalysisofdendriticcell
conventionaldendriticcellprogenitorsinmousebonemarrow.Nat developmentandhomeostasis.Science(NewYork,NY2009,324(5925):392–397.
Immunol2007,8(11):1207–1216. 35. SchlennerSM,MadanV,BuschK,TietzA,LaufleC,CostaC,BlumC,Fehling
13. DolenceJJ,GwinK,FrankE,MedinaKL:ThresholdlevelsofFlt3-ligandare HJ,RodewaldHR:FatemappingrevealsseparateoriginsofTcellsand
requiredforthegenerationandsurvivaloflymphoidprogenitorsand myeloidlineagesinthethymus.Immunity2010,32(3):426–436.
B-cellprecursors.EurJImmunol2011,41(2):324–334. 36. SitnickaE,BrakebuschC,MartenssonIL,SvenssonM,AgaceWW,Sigvardsson
M,Buza-VidasN,BryderD,CilioCM,AhleniusH,etal:Complementary
14. SitnickaE,BryderD,Theilgaard-MonchK,Buza-VidasN,AdolfssonJ,
signalingthroughflt3andinterleukin-7receptoralphaisindispensablefor
JacobsenSE:Keyroleofflt3ligandinregulationofthecommon
fetalandadultBcellgenesis.JExpMed2003,198(10):1495–1506.
lymphoidprogenitorbutnotinmaintenanceofthehematopoieticstem
cellpool.Immunity2002,17(4):463–472. 37. VosshenrichCA,RansonT,SamsonSI,CorcuffE,ColucciF,RosmarakiEE,Di
SantoJP:Rolesforcommoncytokinereceptorgamma-chain-dependent
15. SoCW,KarsunkyH,PassegueE,CozzioA,WeissmanIL,ClearyML:MLL-
cytokinesinthegeneration,differentiation,andmaturationofNKcell
GAS7transformsmultipotenthematopoieticprogenitorsandinduces
mixedlineageleukemiasinmice.CancerCell2003,3(2):161–171. precursorsandperipheralNKcellsinvivo.JImmunol2005,174(3):1213–1221.
38. KikuchiK,KasaiH,WatanabeA,LaiAY,KondoM:IL-7specifiesBcellfateat
16. CarottaS,PangSH,NuttSL,BelzGT:IdentificationoftheearliestNK-cell
precursorinthemouseBM.Blood2011,117(20):5449–5452. thecommonlymphoidprogenitortopre-proBtransitionstageby
maintainingearlyBcellfactorexpression.JImmunol2008,181(1):383–392.
17. FathmanJW,BhattacharyaD,InlayMA,SeitaJ,KarsunkyH,WeissmanIL:
39. DiasS,SilvaHJr,CumanoA,VieiraP:Interleukin-7isnecessaryto
Identificationoftheearliestnaturalkillercell-committedprogenitorin
murinebonemarrow.Blood2011,118(20):5439–5447. maintaintheBcellpotentialincommonlymphoidprogenitors.JExp
Med2005,201(6):971–979.
18. PuzanovIJ,BennettM,KumarV:IL-15cansubstituteforthemarrow
40. PongubalaJM,NorthrupDL,LanckiDW,MedinaKL,TreiberT,BertolinoE,
microenvironmentinthedifferentiationofnaturalkillercells.JImmunol
1996,157(10):4282–4285. ThomasM,GrosschedlR,AllmanD,SinghH:TranscriptionfactorEBF
restrictsalternativelineageoptionsandpromotesBcellfate
19. ChengM,CharoudehHN,BrodinP,TangY,LakshmikanthT,HoglundP, commitmentindependentlyofPax5.NatImmunol2008,9(2):203–215.
JacobsenSE,SitnickaE:Distinctandoverlappingpatternsofcytokine
41. ThalMA,CarvalhoTL,HeT,KimHG,GaoH,HagmanJ,KlugCA:Ebf1-
regulationofthymicandbonemarrow-derivedNKcelldevelopment.
JImmunol2009,182(3):1460–1468. mtheedBiacteedllldinoewang-er.ePgruolcatNioantloAfcaIdd2SacinUdSIAd320i0s9e,s1s0e6n(t2ia):l55fo2r–s5p5e7.cificationof
20. SatheP,VremecD,WuL,CorcoranL,ShortmanK:Convergent
42. BoosMD,YokotaY,EberlG,KeeBL:Maturenaturalkillercelland
differentiation:myeloidandlymphoidpathwaystomurineplasmacytoid
lymphoidtissue-inducingcelldevelopmentrequiresId2-mediated
dendriticcells.Blood2013,121(1):11–19. suppressionofEproteinactivity.JExpMed2007,204(5):1119–1130.
21. WaskowC,LiuK,Darrasse-JezeG,GuermonprezP,GinhouxF,MeradM,
43. IkawaT,FujimotoS,KawamotoH,KatsuraY,YokotaY:Commitmentto
ShengeliaT,YaoK,NussenzweigM:ThereceptortyrosinekinaseFlt3is
naturalkillercellsrequiresthehelix-loop-helixinhibitorId2.ProcNatl
requiredfordendriticcelldevelopmentinperipherallymphoidtissues. AcadSciUSA2001,98(9):5164–5169.
NatImmunol2008,9(6):676–683.
44. YokotaY,MansouriA,MoriS,SugawaraS,AdachiS,NishikawaS,GrussP:
22. CarottaS,DakicA,D'AmicoA,PangSH,GreigKT,NuttSL,WuL:The
Developmentofperipherallymphoidorgansandnaturalkillercellsdepends
transcriptionfactorPU.1controlsdendriticcelldevelopmentandFlt3 onthehelix-loop-helixinhibitorId2.Nature1999,397(6721):702–706.
cytokinereceptorexpressioninadose-dependentmanner.Immunity
45. HolmesML,CarottaS,CorcoranLM,NuttSL:RepressionofFlt3byPax5is
2010,32(5):628–641. crucialforB-celllineagecommitment.GenesDev2006,20(8):933–938.
23. HuangY,SitwalaK,BronsteinJ,SandersD,DandekarM,CollinsC,Robertson
46. LaiAY,LinSM,KondoM:HeterogeneityofFlt3-expressingmultipotent
G,MacDonaldJ,CezardT,BilenkyM,etal:Identificationand progenitorsinmousebonemarrow.JImmunol2005,175(8):5016–5023.
characterizationofHoxa9bindingsitesinhematopoieticcells.Blood
2012,119(2):388–398.
doi:10.1186/1471-2172-14-5
24. InlayMA,BhattacharyaD,SahooD,SerwoldT,SeitaJ,KarsunkyH,Plevritis
Citethisarticleas:Gwinetal.:DifferentialrequirementforHoxa9inthe
SK,DillDL,WeissmanIL:Ly6dmarkstheearlieststageofB-cell
developmentanddifferentiationofB,NK,andDC-lineagecellsfrom
specificationandidentifiesthebranchpointbetweenB-cellandT-cell Flt3+multipotentialprogenitors.BMCImmunology201314:5.
development.GenesDev2009,23(20):2376–2381.