Reconstructing ancestral gene orders with duplications guided by synteny level genome ... PDF
Preview Reconstructing ancestral gene orders with duplications guided by synteny level genome ...
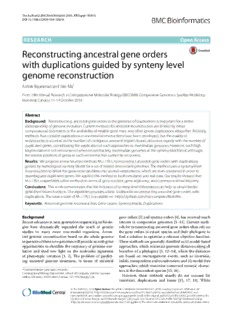
TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 DOI10.1186/s12859-016-1262-8 RESEARCH OpenAccess Reconstructing ancestral gene orders with duplications guided by synteny level genome reconstruction AshokRajaramanandJianMa* From14thAnnualResearchinComputationalMolecularBiology(RECOMB)ComparativeGenomicsSatelliteWorkshop Montreal,Canada.11-14October2016 Abstract Background: Reconstructing ancestralgeneordersinthepresenceofduplicationsisimportantforabetter understandingofgenomeevolution.Currentmethodsforancestralreconstructionarelimitedbyeither computationalconstraintsortheavailabilityofreliablegenetrees,andoftenignoreduplicationsaltogether.Recently, methodsthatconsiderduplicationsinancestralreconstructionshavebeendeveloped,butthequalityof reconstruction,countedasthenumberofcontiguousancestralregionsfound,decreasesrapidlywiththenumberof duplicatedgenes,complicatingtheapplicationofsuchapproachestomammaliangenomes.However,suchhigh fragmentationisnotencounteredwhenreconstructingmammaliangenomesatthesynteny-blocklevel,although therelativepositionsofgenesinsuchreconstructioncannotberecovered. Results: Weproposeanewheuristicmethod,MULTIRES,toreconstructancestralgeneorderswithduplications guidedbyhomologoussyntenyblocksforasetofrelateddescendantgenomes.Themethodusesasynteny-level reconstructiontobreakthegene-orderproblemintoseveralsubproblems,whicharethencombinedinorderto disambiguateduplicatedgenes.Weappliedthismethodtobothsimulatedandrealdata.Ourresultsshowedthat MULTIRESoutperformsothermethodsintermsofgenecontent,geneadjacency,andcommonintervalrecovery. Conclusions: Thisworkdemonstratesthattheinclusionofsynteny-levelinformationcanhelpusobtainbetter gene-levelreconstructions.Ouralgorithmprovidesabasictoolboxforreconstructingancestralgeneorderswith duplications.ThesourcecodeofMULTIRESisavailableon https://github.com/ma-compbio/MultiRes. Keywords: Ancestralgenomereconstruction,Geneorders,Syntenyblocks,Duplications Background geneorders[3]andsyntenyorders[4],hasreceivedmuch Recentadvancesinnext-generationsequencingtechnolo- interest in comparative genomics [5–11]. Current meth- gies have dramatically expanded the reach of genetic odsforreconstructingancestralgeneordersoftenrelyon studies to many more non-model organisms. Ances- the gene orders in extant species and their phylogeny to tral genome reconstruction based on the whole genome find a solution to optimize a relevant objective function. sequencesofthesenewgenomeswillprovideuswithgreat Thesemethodsaregenerallyclassifiedas(i)model-based opportunities to elucidate the trajectory of genome evo- approaches,whichminimizegenomicdistancesalongall lution and shed new light on the molecular signatures branches of a phylogeny [5, 12–14], where the distances of phenotypic variation [1, 2]. The problem of predict- are based on rearrangement events, such as inversion, ing ancestral genome structures, in terms of ancestral indels,transpositionandtranslocation;and(ii)model-free approaches,whichmaximizeconservedsynteniccharac- *Correspondence:[email protected] tersinthedescendantspecies[15,16]. ComputationalBiologyDepartment,SchoolofComputerScience,Carnegie However, these methods usually do not account for MellonUniversity,5000ForbesAvenue,15213Pittsburgh,USA insertions, duplications and losses [15, 17, 18]. While ©TheAuthor(s).2016OpenAccessThisarticleisdistributedunderthetermsoftheCreativeCommonsAttribution4.0 InternationalLicense(http://creativecommons.org/licenses/by/4.0/),whichpermitsunrestricteduse,distribution,and reproductioninanymedium,providedyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktothe CreativeCommonslicense,andindicateifchangesweremade.TheCreativeCommonsPublicDomainDedicationwaiver (http://creativecommons.org/publicdomain/zero/1.0/)appliestothedatamadeavailableinthisarticle,unlessotherwisestated. TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page202of282 progress has been made to incorporate insertions and 2. Asetofancestralsyntenyblocksontheextant deletions [19], efficient reconstruction of gene orders species.Theseblockscapturegenomicregionsacross withduplicationsremainsalargelyopenproblem.There differentgenomeswithhighsequencesimilarity,and existmaximumlikelihoodmethodsthatalsoreconstruct canbedefinedbycomparingmultiplegenomes[15,26]. gene orders with duplications [20], as well as methods Itisassumedthatallhomologousextantsynteny whichutilizereconciledgenetreesintothereconstruction blocksevolvedfromasingleancestralregion[27]. framework[21],butobtainingrobustgenetreesitselfisa 3. Extantgeneorders,withgenesgroupedinto complicatedproblem[22].Anumberofstudiesalsoshow homologousgenefamiliesconsistingoforthologous thatincorporatingduplicationsincurrentreconstruction andparalogousgenesinallspecies. models renders the related optimization problems com- putationallyintractable[23,24]. Our aim is to reconstruct the gene order at a given Reconstructing the ancestral genomes [15, 25] as an ancestorofinterest. Thechallengehere is twofold:given orderingofsyntenyblocksdefinedthroughwholegenome twohomologousgenes,weneedtodistinguishwherethey alignment of the extant genomes [26] can create a more appearontheancestralgenome,andthegeneorderneeds contiguous genome structure. The length of such blocks tobe‘consistent’withtheancestralsyntenyblockorder.In can be controlled, and is typically defined to be greater this paper,we define consistency as finding a gene order than 100 kb. At this resolution, it is common to assume suchthat,foreachconsecutivesubsequenceW ofsynteny that synteny blocks appear at most once in a descen- blocks and gaps between the blocks which is inferred to dant species for amniotes [27], and at most once in the exist in the ancestral genome, there exists a correspond- ancestral reconstruction. These reconstructions usually ingconsecutivesubsequenceSinthegeneordersuchthat have low fragmentation (MGRA [17], for example, pro- the genes and adjacencies in S are preserved within W, ducesexactlyasmanyfragmentsasthemaximumnumber accordingtosomeparsimonycriterionwhichwespecify ofextantchromosomes).However,micro-rearrangements later.Wewanttofindthelargestweightsetofgeneadja- occurring within each synteny block [4] are hidden, pre- cencies which is (i) consistent, with weights defined by venting us from obtaining a comprehensive view of the the status of their phylogenetic conservation [15], while genomeevolutionatthislevel. ensuringthat(ii)thenumberofcopiesofeachgeneinthe In this paper, we propose a new heuristic framework, orderisupperboundedbyaprecomputedancestralcopy MULTIRES,thatintegratesinformationfrommultipleres- number.MULTIRESispresentedasaheuristicthataimsto olutionstoreconstructtheancestralgenome.Themethod achieveboth. uses reconstructed synteny block orders of an ances- The outline of MULTIRES is presented as a flowchart tor to infer gene orders while incorporating duplica- inFig.1.Wefirstinferanancestralorderforthesynteny tions. MULTIRES uses an approach described in [24] for blocks. We use ANGES [29] to find an ancestral recon- finding circular chromosomes in the presence of dupli- struction using the synteny blocks and the species tree cations. We develop a novel method for partitioning as inputs. Note that it is possible to use different meth- families of homologous genes using the synteny blocks ods for this purpose. We used ANGES since, at the time that they occur in. We show that MULTIRES recovers of the experiments, it was one of the few software that up to 18 % more ancestral adjacencies that are missed couldconsidernon-unique,non-universalsyntenyblocks by a method which uses the same optimization rou- intheextantspeciesandproduceanancestorwithatmost tinewithoutusingsyntenyblocks(originallyimplemented a single copy of each block. Since then, we also have the for scaffolding ancestral contigs in [28]) on simulated option of using other methods, such as the new version data, and provide a more comprehensive reconstruction of MGRA [19], but the results were identical to those of of the X-chromosome of the primate-rodent common ANGES on the X-chromosome data set and simulations ancestor. in our experiments. We use the set of contiguous ances- tralregions(CARs),sequencesofsyntenyblocks,obtained fromANGESasaninput. Method Weassumethatwearegiventhefollowingpiecesofdata ThemainideaofMULTIRESissummarizedinFig.1,and alongwithsomeaccompanyingnotation,ispresentedasa asinput. schematicviewinFig.2.Weusethemappingofgenefam- 1. Aresolved(binary)phylogenetictreeonasetof iliesintosyntenyblocksontheextantspeciestopartition extantspecies,andamarkedancestralnodeatwhich a gene family into one or more subfamilies, called local- wewanttoreconstructthegenome.Wearealso izations,whichareexpectedtobesufficientlyfarapartin givenbranchlengthsonthetree.Intheabsenceof the ancestral reconstruction. We construct an ancestral branchlengths,wemayassumethateachbranchhas adjacencygraphonthesetoflocalizations,usingthesetof length1. adjacenciesbetweenfamilieswhichareconservedwithin TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page203of282 Fig.1MULTIRESflowchart.Ahigh-leveloverviewoftheMULTIRESpipeline consecutivesubsequencesintheCARs.Alladjacenciesin genomes. A gene family is a set of genes, either within a thisgrapharegivenaweightbasedontheirconservation singlespeciesoracrossasetofextantspecies,whichare patterninthespeciestree,asdefinedin[15].Wethenuse inferred to have evolved from a single original gene in the algorithm presented by Manˇuch et al. in [24] to find some ancestral species. The set of gene families forms a a maximum weight set of adjacencies such that nodes of partitionofthesetofgenes. specifiedsubgraphscanbearrangedintoasetofcircular Each gene family can be partitioned into a head and a sequences,withconstraintsonhowmanytimeseachnode tail,followingtheusualmethodofdoubling [12,31].The mayappearoverallsequences.Tothebestofourknowl- headandthetailofagenefamilyarereferredtoasmark- edge, this is the only known polynomial time algorithm ers, and there is a one-to-one relation between a given whichoutputsanoptimumweightsetofadjacencieswith head/tail marker and the associated gene family. Thus, a asetofchromosomeswithduplicationsasinput.Finally, chromosomecanbethoughtofasasequenceofmarkers, wecombinetheresultsforallsubgraphstoobtainalinear notnecessarilyunique,withlength2n(nbeingthenum- geneorderforeachCAR. berofgenesinthechromosome,withauniquepairingof In the interest of brevity, we only present the frame- themarkersinpositions2k−1and2k,k ∈{1,..., n}).Two work after the ancestral reconstruction at the synteny successive markers in the sequence that are not paired block level. The inputs we use are the phylogenetic tree, to each other are said to form an adjacency. It is possi- extant gene orders, a set of adjacencies between synteny ble for two markers corresponding to extremities of the blocks,ancestralgenecopynumbers,andasetofCARs. samegenefamilytonotbepairedtoeachother;thesecor- Fordetailsoftheprocessusedtoobtaintheinput,seethe respond to tandem duplications. Given(cid:2)mark(cid:3)ers g,h, an Additionalfile1.Wealsoreferthereadersto[16,30]for adjacency between them is denoted by g,h . However, reference on how to compute conserved characters and forthepurposesofexposition,wewillrepresentgenesin ancestralcopynumbers. examplesandfiguresasasinglesolidelementinsteadofa combinationof2pointmarkers. Definitions Given a gene family g, or equivalently a marker g, an Wefirstintroducesomecommonterminology.Agene,in occurrence of g on an extant genome denotes a specific thecontextofthismanuscript,isashortcontiguousseg- locusatwhichagenebelongingtothisgenefamily/marker ment of a genome. For a given set of extant species, we occurs.Eachgenefamilyandthecorrespondingmarkers assume that we know the exact order of genes in their are associated to a precomputed ancestral copy number TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page204of282 Fig.2aEstimatinglocalizations.HereitshowshowMULTIRESdefinesandinferslocalizations.Colouredsolidwedgesrepresentgenefamilies,with wedgesofthesamecolourbelongingtothesamefamily.Syntenyblocksareindicatedbyhollowwedges,withcolourindicatinghomology.The orientationofthewedgesrepresenttheorientationofthegenes/blocks.Inferringadjacenciesbetweengenefamiliesparsimoniouslyresultsinthe ancestraladjacencygraphshownonthetopleft,withedgesrepresentingadjacenciesbetweengeneends.Wealsohaveasetofcontiguous ancestralregions(CARs)reconstructedattheancestor,eachofwhichconsistsofanorderingoftheancestralsyntenyblocks.Ontherightofthetree, wedisplayanexampleofaCAR,andtheCARafterallsyntenyblockshavebeendoubledintoheadandtailextremities.Wedefinewindowsof length3asconsecutivesubsequencesof3extremitiesontheCAR.Thewindows,indicatedbycolouredlinesegmentsinthefigure,areusedto partitiontheCAR.Inthediagram,weobserveafterpartitioningthatonecopyofthebrowngene(g1)alwaysoccursintheredwindow,andone alwaysoccursinthebluewindow,andneverintheirintersection.Thisallowsustopartitionthebrowngenefamilyintotwosubfamilies,g1andg1, 1 2 calledlocalizations,whicharerestrictedtoappearonlyintherelevantblocks,leadingtothelocalizedadjacencygraphatthebottom.bOptimization andconsensus.Hereweshowalocalizedadjacencygraph(top)withcopynumbersassociatedtoeachlocalization(numbersunderthegenes). PartitioningtheancestralCARsintosegments(blacklinesegments)definesanorderedsequenceofinducedsubgraphs.Usingthealgorithmgiven by[24]oneachinducedsubgraphresultsinasetofadjacenciesshownateachlayer,witheachlocalizationadjacenttoatmostasmanyadjacencies asitscopynumber.Forexample,thebrownlocalizationg1canhaveatmost1copyinthegeneorder,makingitadjacenttoatmost2other 2 localizations.Thealgorithmindicatesthattheseadjacenciesaretothered(g3)andorange(g7)localizations.Finally,wecombinethesubgraphsand 1 1 findalineargeneorderbyfindingthemostfrequentlyconservedadjacenciesandusingtheorderofthesegments.Intheexample,sincethepurple localizationg2isonlyconservedinSegment3,whilethecyanlocalizationg5isconservedinSegment2aswell,wecanresolvethegeneorder 2 1 aroundtheduplicatedorangelocalizationg7 1 [30].Thisnumberdefinesanupperlimitonthenumber regions. The output of general ancestral reconstruction ofcopiesofthegeneintheancestor. techniques is a set of CARs, describing reconstructed Synteny blocks can also be doubled, and give rise to contiguousgenomicsegmentsintheancestor. two extremities. As in the case of markers, a chromo- Cons(cid:2)ider(cid:3)two markers g and h. We say that an adja- somecanalsobedefinedasasequenceofextremitiesof cency g,h is parsimoniously conserved(or equivalently, length2n,withauniquepairingofextremitiesinpositions conserved)intheancestorif,fortwoextantspeciesS and i 2k−1and2k,k ∈ {1,..., n},toformsyntenyblocks.We S in the phylogenetic tree, we find that the two mark- j use the term region to denote a pair of two extremities. ersg andhareadjacentinbothspecies,andtheancestor Thus,aregionrepresentseitherapairofextremitiesfrom underconsiderationliesontheevolutionarypathbetween the same synteny block, or the pair of extremities which thesetwospeciesinthespeciestree.Thetermconserved frameagapbetweentwoadjacentsyntenyblocks.Wewill is also used to refer to a region [a,b] that occurs in two use the notation [a,b] to denote a region, where a,b are extant species such that the ancestor of interest lies on block extremities. From now on, we will only work with theirevolutionarypathinthespeciestree. extremitiesandregionsinordertotakeintoaccountthe An ancestral adjacency graph (or adjacency graph) on orientationoftheblocks. thesetofmarkersisagraphG = (V,E),wherethever- A contiguous ancestral region (CAR) C = c0.....c2k−1 tex set is the set of all markers under consideration, and of length 2k is a sequence of 2k extremities c, such that thesetofedgesistheunionofthesetofparsimoniously i eachregion[c2i,c2i+1]isasyntenyblock,andeachregion conservedadjacenciesbetweenmarkersonagivenspecies [c2i−1,c2i] is an adjacency between synteny blocks. By treeandthesetofedgesbetweentheheadandtailmarkers extension,itcanalsobedescribedasasequenceof2k−1 ofthesamegene[15].Itiseasytoseethatagenomecan TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page205of282 bedescribedasasetofwalksontheadjacencygraph,with Algorithm 1 Algorithm for constructing the adjacency eachwalkalternatingbetweentheconservedadjacencies graph. Copy numbers and adjacency conservation are andtheedgesbetweenheadandtailmarkersofthesame determinedbyparsimony gene [23, 24]. We use a function μ: V → N, called the Input Ancestral CARs, gene family set V , gene con- g ancestral copy number or multiplicity function (see [30] tainmentsinregions,extantregionsandgeneorders, orconsulttheAdditionalfile1fordetailsonhowtoinfer speciestreeT andwindowlength(cid:2). thefunction).Thisspecifiesanupperlimitonthenumber Output An ancestral adjacency graph G = (V,E), copy ofcopiesofasinglemarker(andbyextension,ofasingle numbersμ: V →N gene)allowedinsetofwalks[23,24].Wealsohaveaposi- 1: forallg ∈Vg do tiveweightfunctionw: E→R,whichisinferredfromthe 2: Setyg =−1 phylogeneticconservationofeachconservededge[15]. 3: endfor 4: V ←∅,E←∅ Estimatingextantcontainments 5: forall CARsC = c0.c1...ck−1,ci beingextremities. Given a set of extremities, we can find the sequence of do genesthatarecontainedwithintheseextremitiesinevery 6: forall(cid:2)con(cid:3)secutivewindowsWi =ci...ci+(cid:2)−1do extantspecies.Foragenefamilyg withanoccurrenceof 7: if( g,h (cid:4)arecont(cid:5)ainedandadjacentin length (cid:2) in a given extant species, we say that g is con- region cj,cj+1 (cid:4) (cid:5) tained within an extant region [a,b] in the same species forspeciesS0,andinregion cj(cid:8),cj(cid:8)+1 for if at least half the length of the occurrence (i.e., (cid:2)/2) lies speciesS , 1 withintheregion[a,b].Formally,ifg hashead/tailmark- i≤j,j(cid:8) <i+(cid:2)−1,andtheancestorlieson ers at loci x < y in a given extant species, we say that theevolutionarypathbetweenS andS 0 1 thegenefamilyg iscontainedwithinaregion[a,b]inthe inT)the(cid:4)n (cid:5) (cid:4) (cid:5) species,if,giventhattheextremitiesoftheregionarecon- 8: ifboth cj,cj+1 , cj(cid:8),cj(cid:8)+1 werenot secutiveontheextantgenome,andlocatedatlocila < lb spannedbeforeWi on a given chromosome, one of the following conditions 9: yg ←yg +1,yh ←yh+1. holds. 10: endif (cid:2) (cid:3) 11: V ←V ∪(cid:2) gyg,hyh(cid:3) . 1. la ≤x<y≤lb,or 12: E←E∪ gy(cid:6)g,hy(cid:7)h .(cid:6) (cid:7) 2. x<la <yand|la−y|≥(cid:2)/2or, 13: Computeμ gyg ,μ hyh usingparsimony 3. x<lb <yand|lb−x|≥(cid:2)/2. onT consideringallregionsinW. i Similarly,iftheheadandtailmarkersofgarelocatedat 14: endif locix > y,theng issaidtobecontainedinaregion[a,b] 15: endfor in the extant species if the symmetrical conditions hold. 16: endfor For each extant region, in each species that this region 17: returnG=(V,E),μ. is found in, we thus obtain the sequence of gene family occurrences in this region, if any. Ideally, the gene fam- ily sequence within a region would be conserved across Inferringconservedadjacencies all extant species, and the mapping of these gene fam- Given an ancestral synteny order reconstruction in the ily sequences to the synteny-level reconstruction should form of CARs, a consecutive subsequence of regions defineanancestralgeneorder.However,thisisrarelythe in this reconstruction should inform us about the gene case in real data due to rearrangements, insertions, and content in the corresponding region. To formalize this deletionsatthegenelevelwithintheregions.Thesubse- intuition,wedefineawindowasfollows. quentsectionsaddresshowtofindanorderofthegenes (cid:6) (cid:7) such that the gene content within successive sequences Definition 1 Let C = c0,..., ck−1 be a CAR, where of regions is preserved, and the total weight of adja- each c is an extremity. A window of length (cid:2) on C is i cencies between the genes in a given sequence, inferred (cid:4)a conse(cid:5)cutive subsequence W = ci...ci+(cid:2)−1, with each phylogenetically,ismaximized. cj,cj+1 ,i≤j<i+(cid:2)−1,beingaregio(cid:4)ninC.(cid:5)Theinteger(cid:2) iscalledthewindowlength.Aregion cj,cj+1 issaidtobe FindinggeneordersinaCAR spannedbythewindowW ifcjandcj+1areadjacentinW. We now use the extant gene sequences in regions, con- servedmarkeradjacenciesandcopynumbers,thespecies Fig.2ashows2differentwindowsoflength3definedon tree,andCARsastheinputtofindputativeancestralgene theancestralCAR,andtheconstituentregionsaslocated familyorders. in a set of extant species. We use windows to partition TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page206of282 genefamilies(andbyassociation,markers)intosubfami- v,wecandefinethe“inversemap”ψ−1([a,b])astheset lieswhichareexpectedtooccur‘farapart’intheancestral oflocalizationsassociatedtotheregion[a,b]. genome.Formally,wehavetheconceptoflocalizations. Optimizingwithinasegment Definition 2 Let g be a gene family, and let W = Given the adjacency graph G on the set of localizations, {W :0≤ i<k}beasubsetofallwindowsoflength(cid:2)inall andasetofassociationsoftheselocalizationstoregions, i CARs,suchthat∀W ∈Wthereexistregionsr,r(cid:8)spanned we can use the linear structure of the CARs to design a i byW,whichcontaing inextantspeciesS andS respec- local optimization scheme. In order to do this, we again i a b tively,suchthatr,r(cid:8) arenotspannedbyanyotherwindow consider consecutive subsequences of L extremities, or inW,andtheancestorofinterestliesontheevolutionary equivalently L−1 regions, on the CARs, and try to find pathbetweenS andS .Notethatrandr(cid:8)maybethesame local gene orders in each of these subsequences. These a b region. subsequences,whichwewillcallsegments,arethussimilar Alocalizationofg(andbyextension,ofthemarkersofg), towindows,exceptthattheuser-definedparameterspec- isasubfamilyg ofg definedsuchthatalladjacenciestog ifying their length L is required to be at least as long as i fromothergenefamiliesconservedwithinthewindowW thewindowlength(cid:2)usedtoconstructG.Therefore,aseg- i areadjacentonlytog. mentmaycontainmanywindows,andbyextensionmany i localizations of the same original gene family. Figure 2b showshowsegmentsaredefined. Inotherwords,ifanoccurrenceofginaregion[a,b]in Once we have a set of segments of the CARs, we find someextantspeciesisalwaysadjacenttoamarkerp,and subgraphsinGrestrictedtothesetofregionsspannedby anoccurrenceofginadifferentregionwhichisnotinthe eachsegmentandfinda‘good’setofadjacenciesineach same window as [a,b] is adjacent to a distinct marker q, subgraphasfollows. thenwecanpartitionthegenefamilyofgintooccurrences adjacenttopandoccurrencesadjacenttoq. 1. ForeachsegmentoflengthLonaCAR,wherethe adAjalcgeonrcitihems b1etdweesecnribtehsemho.wAstooduetpfiunte, lwoecaolibzatatiionnpsaarntid- correspondingL−1regions(cid:8)are{[bi,bi+1]}k≤i<k+L, findtheinducedsubgraphG ofGonthefollowing tions of the gene families (markers) into localizations, setoflocalizations, whichwedenotebyV,asetofparsimoniouslyconserved (cid:8) afudnjaccteionnciμes: Vbet→weNen,wlohcicahlizaastsiiognnss,adceonpoytendumbybeEr,toaneadcha V(cid:8) = ψ−1([bi,bi+1]), k≤i<k+L localization using the parsimony algorithm detailed in [30].ThesetsV andEareusedtodefinealocalizedadja- i.e.,thesetoflocalizationsassociatedwithregionsin cency graph G = (V,E), which differs from the original thisse(cid:6)gm(cid:7)ent. adjacency graph on the set of markers in that the ver- 2. Letμ G(cid:8) betherestrictionofthecopyn(cid:6)umber(cid:6) (cid:7)(cid:7) ticesarenowlocalizations.Thealgorithmissummarized functionμtothelocalizationsinV(cid:8).Use G(cid:8),μ G(cid:8) inFig.2a,whichshowsthelocationsofthewindowsand astheinputtoManˇuchetal.’s[24]algorithmtofind their constituent regions in the extant species, as well as (cid:8) amaximumweightsetofadjacenciesinG which adescriptionofhowtheadjacencygraphonlocalizations willadmitasetoflinearorcircularchromosomes. differsfromtheadjacencygraphonmarkers. Thealgorithmfindsasetofadjacenciesinwhich Thealgorithmupdatesthecopynumberfunctionμso eachlocalizationv∈V(cid:8)isadjacenttoatmostμ(v) that the total copy number of all localizations of a given otherlocalizations. genefamily is equalto the originalestimatedcopy num- 3. Returnthesetofalladjacenciesfoundwithineach berofthegenefamily.Thisconstraintisenforcedbythe segment. following heuristic: (i) delete localizations which are not (cid:8) involved in any adjacencies, and (ii) decrease the copy For subgraph G, we obtain a maximum-weight set of numberofthelocalizationwiththehighestcopynumber adjacenciesbetweenlocalizationssuchthat(i)eachlocal- iterativelytilltheconditionissatisfied.Thealgorithmalso ization v is adjacent to at most μ(v) other localizations, (cid:8) associates each localization to a set of regions in which and(ii)thereisasetoflinear/circularwalksinG which they could be contained in the ancestor. This is repre- uses exactly this set of edges. The set of adjacencies will sented as a map ψ: V → 2R, whereV is the set of be similar for consecutive segments, with one possibly localizations,andRisthesetofallregions,suchthatfor extendingtheother.Thedefinitionsofthesegmentsand anylocalizationv ∈ V ofamarkerv,ψ(v )isthesetof theresultontheassociatedsubgraphsafteroptimization k k regionsinwhichvcanbefoundinsomeextantspecies,as illustratedinFig.2b. observedinLine7ofthealgorithm.Sinceeachregionin Thepreviousstepisthebottleneckintheprocess,con- ψ(v )canonlybeassociatedwithasinglelocalizationof sisting of multiple maximum matching routines, which k TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page207of282 (cid:6) (cid:7) take roughly O |E(cid:8)|3/2 each, where E(cid:8) is the set of adja- The final order is returned as a set of concatenated cencies between localizations in a subgraph. However, if paths, with the order and orientation of each path thewindowandsegmentlengthsarecarefullychosen,this expectedtobeindicativeoftherelativeorderofthemark- step can be completed under 400s for an instance with ers compared to the neighbouring paths, as shown in ∼730genesintheextantspecies,comparedtoover1000s Fig.2b. forotherparametercombinationsonasingleIntelXeon 2.20 GHz processor, while, as shown in the Additional Results file1,varyingtheparametersdoesnotsignificantlyaffect We used MULTIRES to reconstruct the X-chromosome thereconstructionquality. gene order of the primate-rodent ancestor for both sim- ulated data as well as real data. We used human, chim- Constructingthefinalordering panzee, rhesus macaque, marmoset, rat and mouse as The final step of the method is to find a consensus ingroups, and dog, cattle, pig and horse as outgroups. sequenceofmarkersusingadjacencieskeptforeachseg- The genes and species tree were obtainedfrom Ensembl mentofeachCAR.Wemergetheadjacencieskeptineach (the species tree is illustrated in Fig. 3). We used syn- segment to create an adjacency graph for a single CAR. tenyblocksofresolution100Konthedescendantspecies, Inthisadjacencygraph,thecopynumbersofthelocaliza- computed using whole genome comparison. No synteny tionsareinheritedfromthepreviousstep,butoverlapping blockappearedtwiceinanyspecies,acommonassump- segmentsmayhaveconflicting(cid:2)adja(cid:3)cencies. tionforamniotes[27],buttheycouldbeuniquetoasingle W(cid:6)e(cid:2)ass(cid:3)ig(cid:7)n eac(cid:6)h(cid:2)adj(cid:3)a(cid:7)cenc(cid:6)y(cid:2) x,(cid:3)y(cid:7) a weigh(cid:6)t(cid:2)def(cid:3)in(cid:7)ed by descendant. wgt x,y =P x,y /T x,y ,whereP(cid:2) x,y(cid:3) isthe Unless otherwise mentioned, the method is run with numbe(cid:6)r(cid:2)ofsu(cid:3)(cid:7)bgraphsinwhichtheadjacency x,y iskept, parameters of window length 25 and segment length 65. andT x,y isthenumberofsubgraphsinwhichboth IntheAdditionalfile1,weshowthattheparametersinthe xandyareassociatedtosomeregion,notnecessarilythe rangechosendonotsignificantlychangethequalityofthe same. reconstruction,measuredasthenumberofancestraladja- We then greedily delete the lowest weight adjacency cenciesrecovered.However,longerwindowandsegment suchthatthedegreeoftheadjacentmarkersexceedstheir lengthsleadtoaddedcomplexity,sincetheinducedsub- copy numbers. Repeating this process results in a set graphs grow larger. Our choice of window and segment of adjacencies between localizations which have at most lengths is based on minimizing the running time for the as many adjacencies as their copy numbers. We use the chosenparameters. followingmethodtofindtheorderoflocalizations. Resultsonsimulateddata 1. Rankthelocalizationsbasedonthesequenceof We created 50 simulated data sets each at 2 differ- regionstheyarecontainedin.Forexample,a ent rearrangement, duplication, insertion and deletion localizationiscontainedinasequencec .c .c of rates. The simulation methodology is described in the 0 1 2 extremitiesisrankedhigherthanonethatappearsin Additionalfile1.Thetworearrangementrateswerecho- thesequencec .c .c onthesameCAR. sen so that the number of breakpoints between ingroup 1 2 3 2. Startingatthehighestrankedlocalization,ifithasa species form lower and upper bounds to those found in uniqueneighbour,addtheneighbourtotheexpected therealdata.Wecalledthetwosimulationsetsatdifferent path. 3. Traversethegraphinthedirectionoftheneighbour ofthelastlocalizationaddedtothepath. 4. Ifalocalizationhasmorethan1neighbour,traverse thegraphinthedirectionofthehighestranked neighbour,takingintoaccountthenumberofcopies ofthatneighbourused.Ifallcopiesoftheneighbour havebeenused,movetothenexthighestranked neighbour. 5. Ifthereisatieintheranking,constructthepaths fromthetiedneighboursseparately,andaddthemto thepathsequenceinorderofthehighestranked endingvertexinthepaths. 6. Ifthetraversalreturnsacycle,deletethelastedge Fig.3Mammalianspeciestree.Thespeciestree,withtheancestorof traversed. interestmarkedinred 7. Returnpath(s)obtainedintheorderoftheirtraversal. TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page208of282 rearrangement rates the low rearrangement simulations simulationsets,usingfixedparametervaluesasrecovered andthehighrearrangementsimulationsrespectively. by FPMAG, MGRA2 and MULTIRES. MULTIRES yields a significantly longer reconstruction, of average length Simulationresults (cid:6) (cid:7) ∼627 adjacencies for the low rearrangement sets, with WeranMULTIRESfor 8 parametercombinations,vary- ∼ 78 % true positives, and average length ∼583 for the 2 ing segment lengths and window lengths from 15 to highrearrangementsets,with∼ 70%truepositives.The 85 at intervals of 10. We assessed our results by com- falsepositiverateinbothcasesiswellunder10%.Incom- paring against FPMAG. FPMAG is a method derived parison,FPMAGreturnsareconstructionwithanaverage from FPSAC [28], a tool for scaffolding ancestral bacte- of∼445and∼416adjacencies,withtruepositiveratesof rialcontigs.WhileFPSACandFPMAGarenotintended ∼ 56 % and ∼ 50 % for the two simulation sets respec- for use on mammalian genomes, they use the max- tively,whileMGRA2,whichignoresduplications,findsat imum matching routine described in [24], and the most ∼35 % true positives and about ∼31 % false posi- conceptofrepeatspanningintervals[28]toresolvedupli- tivesinreconstructionsofaveragelength∼496and∼445 cations. To the best of our knowledge, this is the only respectively. publicly available software package that computes an Comparing the fragmentation levels of the methods ancestral reconstruction in the presence of duplications used, we found that FPMAG produces ∼39 CARs (stan- with only gene families and a phylogenetic tree as input. dard deviation = 5.16) in the low rearrangement sim- Since it uses the same optimization routine as MUL- ulations on average, while in the high rearrangement TIRES, we feel this comparison can be used to gauge simulations,itproduces∼45CARs(s.d.=6.85).MGRA2 how the introduction of synteny blocks can augment produces only 1 CAR, but does not recover most of the the reconstruction process. We also compare MULTIRES genecontent,asseeninFig.4.Usingbothsyntenyblocks againstMGRA2[19],inordertohighlighthowthepres- and gene families, MULTIRES finds on average 46 frag- ence of duplications can obfuscate ancestral gene order mentsinthelowrearrangementsimulationset,and∼59 reconstruction. fragmentsinthehighrearrangementsimulationset.How- Weusedthenumberofancestraladjacenciesrecovered ever,thereisatotalorderonthesefragments,whichare in the reconstruction to measure reconstruction quality. linearly ordered on 1 or 2 CARs formed at the synteny Figure 4 compares the true positive, false positive and block level, for both rearrangement rate sets on average. falsenegativeratesofreconstructedadjacenciesforboth Therefore, the reconstructed gene order is reconciled Fig.4Adjacencyrecoverycomparison.Comparisonofadjacencytruepositive(TP),falsepositive(FP)andfalsenegative(FN)ratesforMULTIRES againstFPMAG(cf.[28])andMGRA2[19]onboththelowrearrangementrateandhighrearrangementratesimulations.FPMAGfailstorecovera numberofancestraladjacencies,despiteusingrepeatspanningintervals.TheresultsusingMGRA2areprovidedtocontrasthowmuchofa differencethepresenceofduplicationscanmakeinareconstruction TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page209of282 with the synteny blocks and their adjacencies which are to10)isusuallycompetitivewiththenumberofancestral expectedtocontainthosegenes. adjacenciesrecovered.Indeed,inthehighrearrangement sets, more intervals of length 6 are recovered (≥ 70 %) Largerscaleconservation than adjacencies, as seen by comparing with Fig. 4. This We also examined the number of recovered common showsthatMULTIRESfindssmallneighbourhoodsofco- intervals,asdefinedin[32].Acommonintervaloflength localized genes present in the ancestor, even if the exact 2k > 2 between two genomes is a set of 2k (not neces- geneorderishardtorecover. sarilydistinct)markers,orequivalentlykgenes,whichare Another reason for the loss of large intervals is our found to occur consecutively in both genomes, with the relianceonthesynteny-levelreconstruction.Anyintervals internalorderofthegenesunspecified. whichcontainmarkersfromtwoormoredifferentCARs Here,wedonotcompareagainstMGRA2andFPMAG willbelost.Forexample,ifthesyntenylevelreconstruc- for the following reasons: the gene content recovered by tionproduces2CARsforasingleancestralchromosome, MGRA2iscomparativelylow,whichprecludesthepossi- andoftwoadjacentgenes,eachoneisfoundinaregionon bilityofrecoveringalargenumberofcommonintervals, exactly1CAR,thennointervalscontainingbothofthem andFPMAGhasahighfragmentationrate,duetowhich canberecovered. very few sufficiently long common intervals are recov- ered. In comparison, MULTIRES has the advantage of Resultsonrealdata:ancestralX-chromosomeofthe havingorderedthegenesonancestralCARs,whichallows primate-rodentcommonancestor for a better comparison against the simulated ancestral For the experiments on the real data, we attempted to genome. reconstruct the X-chromosome of the primate-rodent Comparing the number of recovered intervals at both common ancestor. The X-chromosome was chosen due rearrangement rates for fixed parameter values, we to the high concentration of gene duplications (∼20 % obtained Fig. 5. We first point out how the number of of the gene families). We used synteny blocks with reso- commonintervalsdecreasesrapidlywithintervallength. lution 100 Kb, and found a synteny-level reconstruction Thisisaresultofthenumberofgenesintheancestorthat oftheancestorusingANGESconsistingofasingleCAR werenotfoundinthereconstruction:sincesuchgenesare of 374 synteny blocks. The set of extant genes consisted neverfound,allintervalscontainingthemarelost.How- of 626 extant gene families occurring in at least 2 extant ever, the number of short intervals recovered (length 6 species,atleastoneofwhichisaningroupspecies.Gene Fig.5Intervalrecovery.Theratiosofintervalsrecoveredagainstthesizeoftheintervals,forbothsimulationsets.Notethesteadydifferenceinthe ratioofrecoveredintervals:fewerintervalsinthehighrearrangementsetarerecovered.Theredplothasbeenshiftedby0.1alongthex-axisfor easierviewing.Longerintervalsarelostduetothenumberofgeneswhicharenotrecoveredinthereconstruction TheAuthor(s)BMCBioinformatics2016,17(Suppl14):414 Page210of282 families with inferred ancestral content greater than 15 for example, using long reads to improve short-read werediscarded. assembly is a well-studied principle [33]. Till recently, As before, we compared the method against MGRA2 though,ancestralreconstructionreliedongenomicinfor- and FPMAG. To evaluate the reconstruction, we con- mationatasingleresolution.Longerregionswereinferred sider the total number of genes which are recovered in viaancestralconservation[16,28,29,34]. thereconstruction,thenumberofadjacenciesfound,and Thecurrentmethodreliesonthequalityofthesynteny- the number of fragments reconstructed. Table 1 sum- level reconstruction. While this provides the added flex- marizes the results on the real data. We found that a ibility of using a given synteny-level reconstruction, if large proportion of the total possible gene content was the original synteny-level reconstruction is still highly lost. Of 746 possible genes (summing up the ancestral fragmented, we cannot hope to achieve better results at copies of each gene family), we found around 518 genes, the gene-level reconstruction. Using the gene-level data withamaximumof553,andaminimumof478depend- to correct the synteny-level reconstruction would be an ing on the parameters used. Similarly, of 749 conserved interesting next step for the current model. A rigorous ancestral adjacencies, we recovered around 468, with a formalization and analysis of the current model, along minimumof459andamaximumof480.Thisisanadja- withcomparisontoimprovedmodels,couldprovideuse- cency recovery rate of about 62 %. The method found fulinsightsintotherobustnessofthemethodanditsplace 57 linear fragments on average, ordered on the single infuturereconstructionpipelines. CARtoobtainagene-levelrepresentationoftheancestral Inordertovalidatetheresultofthereconstruction,we X-chromosome. usedbothadjacencyandintervalconservationasmetrics. In comparison, MGRA2, which does not take duplica- One of the problems we ran into while computing inter- tions into account, recovered 132 genes, and of the 749 valconservationwastheinabilitytorecoverlargeintervals conserved adjacencies, only recovered 42. It also created due to loss of gene content. In this regard, it would be 88noveladjacencieswhicharenotphylogeneticallysup- useful to consider the concept of approximate common ported.However,MGRA2isabletocontrolthenumberof intervals[35–37].Weaimtoanalyzeintervalconservation fragmentscreatedintheprocess,andfindsonly1CAR. inthiscontextinfuturework. FPMAGfound429genesand350adjacencies,arranged Aswithotherhomologybasedreconstructionmethods, into79CARs.Thisisanadjacencyrecoveryrateofabout MULTIREScannotdetectsignalsofconvergentevolution. 47 %. Both MULTIRES and FPMAG are homology based In Fig. 4, the false positives found by MULTIRES are all methods,anddonotcreateanyunsupportedadjacencies attributable to convergent evolution. In the high rear- outside those conserved. As in the simulations, the false rangement rate simulations, for example, an average of positivescreatedbythesetwomethodscanbeattributed 213 adjacencies, out of a total of ∼885 conserved in the solely to convergent evolution scenarios. However, they descendantspecieswerenotpresentintheancestor.Fur- willalsofailtorecoverancestraladjacencieswhicharelost thermore,10%to15%ofthefalsenegativesareancestral alongallbranchesofthespeciestree. adjacenciesthatwerelostduringevolution.Thisproblem isexacerbatedathigherrearrangementrates. Discussion The use of low-resolution genomic information in order Conclusion to improve the accuracy of high-resolution genomic Theresultspresentedinthismanuscriptprovideaproof reconstructionisnotlimitedtoancestralreconstruction; ofconceptonhowsyntenyblockinformationobtainedvia multiple genome comparison can help ancestral recon- Table1Comparisonofthegeneorderreconstructionofthe struction at a higher resolution where duplications may primate-rodentancestralX-chromosomeusingMGRA2,FPMAG beprevalent.Theimplicationsofthemethodaretwofold: andMULTIRES (i) even with a high level of fragmentation, it is possi- Conserved MGRA2 FPMAG MULTIRES ble to obtain a relative order of the fragments on the Genes 746 132 429 518.12(19.81) synteny-level reconstruction, and (ii) the synteny blocks allow us to disambiguate duplications, which are nor- Adjacencies 749 130 350 468.31(6.05) mally discarded in reconstruction methodologies, thus Recovered - 42 350 468.31(6.05) preventingfragmentationandobtainingamorecomplete Fragments N/A 1 79 53.16(4.76) reconstruction.Fromamethodologicalpointofview,the Therowfortotaladjacenciesindicatesthenumberofadjacenciesfoundinthe method described relies on the decomposition of the reconstruction.Thethirdrowindicatesthenumberofreconstructedadjacencies reconstruction problem into many smaller, overlapping whichareconservedin2ormoredescendantspecies.NotethatFPMAGand MULTIRESonlyrecoverconservedadjacencies.MGRA2canalsolimitthenumberof subproblems,whichtoourknowledgeisanoveltechnique CARsreconstructedandfindasingleCAR.TheresultsforMULTIRESareaveraged in ancestral reconstruction. The use of the maximum overallparametercombinations.Thelowstandarddeviationsdemonstratethe robustnessofthemethodtoparameterchoices matchingroutine[24]forthesesubproblemsinsteadofon
Description: