Table Of ContentRESEARCH ARTICLE
Morphologic Evidence for Spatially Clustered
Spines in Apical Dendrites of Monkey
Neocortical Pyramidal Cells
Aniruddha Yadav,1,2 Yuan Z. Gao,1,2 Alfredo Rodriguez,1,2 Dara L. Dickstein,1,2 Susan L. Wearne,1,2†
Jennifer I. Luebke,3 Patrick R. Hof,1,2 and Christina M. Weaver2,4*
1FishbergDepartment of NeuroscienceandFriedman Brain Institute,Mount SinaiSchoolof Medicine,NewYork, NewYork10029
2ComputationalNeurobiologyandImagingCenter,Mount SinaiSchoolof Medicine,NewYork, NewYork10029
3Departmentof AnatomyandNeurobiology,Boston UniversitySchool of Medicine,Boston,Massachusetts 02118
4Departmentof Mathematics,Franklin andMarshallCollege, Lancaster, Pennsylvania17604
ABSTRACT spineson280apicaldendriticbranchesinlayerIIIofthe
The general organization of neocortical connectivity in rhesus monkey prefrontal cortex. By using clustering
rhesus monkey is relatively well understood. However, algorithms and Monte Carlo simulations, we quantify the
mounting evidence points to an organizing principle that probability that the observed extent of clustering does
involves clustered synapses at the level of individual not occur randomly. This provides a measure that
dendrites. Several synaptic plasticity studies have tests for spine clustering on a global scale, whenever
reported cooperative interaction between neighboring high-resolution morphologic data are available. Here we
synapses on a given dendritic branch, which may poten- demonstrate that spine clusters occur significantly more
tially induce synapse clusters. Additionally, theoretical frequently than expected by pure chance and that spine
models have predicted that such cooperativity is advan- clustering is concentrated in apical terminal branches.
tageous, in that it greatly enhances a neuron’s computa- These findings indicate that spine clustering is driven by
tionalrepertoire.However, largelybecauseofthelackof systematic biological processes. We also found that
sufficient morphologic data, the existence of clustered mushroom-shaped and stubby spines are predominant
synapses in neurons on a global scale has never been in clusters on dendritic segments that display prolific
established. The majority of excitatory synapses are clustering, independently supporting a causal link
found within dendritic spines. In this study, we demon- between spine morphology and synaptic clustering.
strate that spine clusters do exist on pyramidal neurons J.Comp.Neurol.520:2888–2902,2012.
by analyzing the three-dimensional locations of (cid:1)40,000 VC 2012WileyPeriodicalsInc.
INDEXING TERMS: clustering; dendritic spines; plasticity; morphology; image analysis
A large body of theoretical and experimental evidence andNevian,2008;Larkumetal.,2009).Importantnewevi-
points to synaptic clustering as a basic organizing princi- dencesuggeststhatgroupsofspatiallylocalizedsynapses
ple of neuronal connectivity. Theoretical models have ona dendrite can process functionally similar information
shownthatpreciselytimedandspatiallylocalizedsynaptic frompresynapticcellassemblies(Takahashietal.,2012).
inputs can sum nonlinearly (Mehta, 2004; Govindarajan It has also been reported that newly formed spines
et al., 2006; Larkum and Nevian, 2008), generating a preferentially grow near synapses activated through
responsethatiseithergreaterorlessthanexpectedifthe
inputs were simply added together. Nonlinear summation
†WededicatethisarticletoSusanL.Wearne,ourfriend,colleague,and
ofsynapticinputsincreasesaneuron’sflexibilitytodiffer- mentor,whopassedawayinSeptember,2009.
entiate spatiotemporal input patterns and greatly enhan- Grant Sponsor: National Institutes of Health; Grant numbers:
MH071818,AG035071,AG025062,andAG00001.
ces its computational efficiency (Poirazi and Mel, 2001;
*CORRESPONDENCETO:ChristinaM.Weaver,DepartmentofMathematics,
Poirazi et al., 2003; Polsky et al., 2004; Gordon et al.,
Franklin and Marshall College, P.O. Box 3003, Lancaster, PA 17604-3003.
2006). Such nonlinear summation in spatially localized E-mail:[email protected]
synapseshasalsobeenobservedexperimentally (Larkum Received August 19, 2011; Revised January 30, 2012; Accepted January
31,2012
DOI10.1002/cne.23070
Published online February 8, 2012 in Wiley Online Library
VC 2012WileyPeriodicals,Inc. (wileyonlinelibrary.com)
2888 TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience 520:2888–2902(2012)
Spineclustering in neurons:morphologicevidence
learning-relatedinductionoflong-termpotentiation(LTP), (Rodriguez et al., 2003, 2006, 2008, 2009; Dumitriu
possiblyleadingtospineclustering(DeRooetal.,2008). et al., 2011) for all automated morphology reconstruc-
Recent experiments have also suggested that the spa- tions included in this study. Still, establishing the exis-
tially colocalized synapses can be regulated simultane- tence and biological significance of clusters of spines is
ously and provide a mechanistic framework that could inherently problematic. Visual inspection may identify
account for the emergence of spatial synapse clusters. clusters but cannot differentiate between clustering
Harvey and Svoboda found that early-phase LTP (E-LTP) resulting from a systematic biological process and clus-
inductionatonesynapseloweredthethresholdforE-LTP teringthatoccurspurelybychance.Asautomatedimage
inductionatsynapseswithina(cid:1)10-lmneighborhoodon analysis methods render high-resolution morphologic
the same dendrite branch (Harvey and Svoboda, 2007). data more accessible, we need a subjective method to
Specifically, Harvey and Svoboda showed that protein- test whether spine clustering occurs more often than
synthesis-independent cross-talk between a synapse and expectedbychance.
itsneighborsexistedafterinductionofE-LTP.Govindarajan Herewepresentanovelapproachthatpairsacluster-
and colleagues (2011) studied the protein-synthesis-de- ing algorithm with Monte Carlo simulations to perform
pendent latephase of LTP (L-LTP) and demonstratedthat suchatest.Weintroduceameasurecalledthe‘‘C-score,’’
theefficiencyofL-LTPinductiononadendriticspineneigh- which is independent of spine density and quantifies the
boringaspineinwhichLTPhadalreadybeeninducedwas probability that the observed number of spine clusters
inversely proportional to the distance between the two occurred randomly. We analyzed the spatial locationand
spines.Furthersupportforclusteredsynapsesarisesfrom shape of spines from 280 dendrite branches of seven
the existence of dendritic spikes, which are likely to be layer III prefrontal cortex (PFC) pyramidal neurons from
evokedbyspatiotemporallylocalizedsynapticinputs(Lar- three adult rhesus monkeys, imaged in their entirety at
kumetal.,2001;Murayamaetal.,2007). highresolution.Weshowthatthefrequencyofspineclus-
Despite these clues, the question of whether synapse tering on apical terminal branches of these neurons is
clusteringoccurs onaglobal scale onbranchesthrough- unlikelytooccurmerelybychance,suggestingthatasys-
out the dendritic arbor is difficult to explore. Several tematic biological process contributes to the observed
network-level approaches are able to identify synaptic clustering. We also found that clusters have higher den-
connections between specific neurons. Recent technolo- sities of mushroom-shaped and stubby spines on den-
gies, including light-activated ion channels (Petreanu drites with prolific clustering than on dendrites with
etal.,2007),glutamateuncaging(Nikolenkoetal.,2007), sparseclustering,indicatingthatspineshapeandcluster-
trans-synaptictracers(Wickershametal.,2007),genetic ingmaybecoregulated.
labeling(Livetetal.,2007),andinvivomultiphotonimag-
ing (Kerr and Denk, 2008), have made great strides in
MATERIALS AND METHODS
improving our understanding of how neurons connect to
form circuits. However, identifying individual synapses Cell imaging, 3-D neuron reconstruction,
participating in these connections and determining and morphometric analysis
whethersynapsesgrouptogetherfunctionallytotransmit Seven layer III pyramidal neurons filled during electro-
related information falls beyond the purview of these physiological recordings in in vitro slices prepared from
technologies. the dorsolateral PFC (area 46) of three young adult male
There is much to be gained by examining the location rhesus monkeys (Macaca mulatta, ages ranging from 5.2
and distribution of synapses on individual neurons. On to6.9years)wereusedinthepresentstudy.Thesemate-
many neurons, dendritic spines provide a morphologic rials were derived from animals killed as part of other
reflection of thepresence ofexcitatorysynapses.Confo- ongoing studies involving a larger set of subjects. These
cal and multiphoton laser scanning microscopy (for three monkeys were part of a cohort used in studies of
review see Wilt et al., 2009) have allowed us to visualize normal aging on the brain. Animals were behaviorally
spines across entire neurons, making analysis of the assessed on a battery of cognitive tasks (Chang et al.,
three-dimensional(3-D)spineshapeandlocationtheoret- 2005; Luebke and Amatrudo, 2010), but no invasive
icallypossible.However,manualanalysisofspinesispro- procedures had been performed on them prior to the
hibitively time consuming and widely subject to operator timeofdeath.
variability. Recent software packages now can analyze The monkeys were housed at the Boston University
dendrite and spine morphology automatically (for review Laboratory Animal Science Center (LASC) in strict
see Lemmens et al., 2010; Meijering, 2010), greatly accordancewithanimalcareguidelinesasoutlinedinthe
reducing manual interventions to initial setup and posta- NIH Guidefor the care and useof laboratory animals and
nalysis editing. In particular, we utilized NeuronStudio theU.S.PublicHealthServicepolicyonhumanecareand
TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience 2889
Yadav et al.
useoflaboratoryanimals.BostonUniversityLASCisfully 2003).Toensurethatthedendritictreewaslargelycom-
accredited by the Association for Assessment and plete,onlycellswithoutcutdendritesintheproximalhalf
Accreditation of Laboratory Animal Care, and all proce- ofthedendritictreeswereused;theywereneverusedif
dureswereapprovedbytheInstitutionalAnimalCareand the apical trunk was cut. The yz-projection in Figure 1a
Use Committee (IACUC). Electrophysiological and cell demonstratesthatmuchoftheapicalarborwascaptured
filling protocols were performed as described in detail successfully.
elsewhere (Luebke and Chang, 2007; Luebke and Thecustom-designedsoftwareNeuronStudio(Rodriguez
Amatrudo,2010). et al., 2006, 2008, 2009) was used for automated
Ketamine(10mg/ml)wasusedtotranquilizethemon- reconstruction of the dendritic arbor and for automated
keys,followingwhichtheyweredeeplyanesthetizedwith identificationandshapeclassificationofspines(Fig.1b–d).
sodiumpentobarbital(toeffect15mg/kg,i.v.).Next,the Spine shape (mushroom, stubby, thin) was determined
monkeys underwent a thoracotomy, and a craniotomy usingcriteriapreviouslydescribed(Rodriguezetal.,2008).
was performed. A biopsy was conducted to obtain Subsequent manual examination and editing reduced the
10-mm-thickblocksofthePFC(area46)containingboth incidenceoffalsepositivesandnegatives.
the upper and lower banks of the principal sulcus. A Eachdendritebranchtraverseda3-Dpathintheimage
vibrating microtome was used to cut 400-lm-thick coro- but was represented here as a one-dimensional (1-D)
nal slices, which were placed in 26(cid:2)C oxygenated (95% segmentwithtotallengthequaltothebranch’straversed
O /5% CO ) Ringer’s solution (concentrations in mM: pathlength.Thelocationofeachspinealongthedendrite
2 2
26 NaHCO , 124 NaCl, 2 KCl, 3 KH PO , 10 glucose, was represented by first projecting the spine head
3 2 4
2.5CaCl ,1.3MgCl ,pH7.4;allchemicalsobtainedfrom onto the dendrite, then measuring the 1-D path length
2 2
Sigma, St. Louis, MO). Slices equilibrated for at least fromthestartoftheparentdendritebranchtothatpoint
1hourandweremaintainedforupto12hours.Thetime (Fig.2a).
betweenperfusionandPFCslicepreparationwasapprox- Because basal dendrites are morphologically distinct
imately 10–15 minutes. For recording, individual slices from apical dendrites and integrate inputs somewhat
werepositionedunderanylonmeshinasubmersion-type differently (Spruston, 2008), we restricted the present
slice-recording chamber (Harvard Apparatus, Holliston, analysis to apical dendrites. Similarly, because evidence
MA) and constantly superfused with 26(cid:2)C, oxygenated intheCA1fieldofthehippocampussuggeststhatspikes
Ringer’ssolutionatarateof2–2.5ml/minute. aregeneratedmoreeasilyinobliqueratherthanterminal
Electrodes were pulled by a horizontal Flaming and branches (Gasparini et al., 2004; Losonczy and Magee,
Brown micropipette puller (model P-87; Sutter Instru- 2006),weconsideredobliqueandterminalbranchessep-
ments,Novato,CA)andfilledwithapotassiummethane- arately. The oblique dendrites are distal to the primary
sulfonate-based internal solution (concentrations, in trunk, correspond to secondary and tertiary levels of
mM): 122 KCH SO , 2 MgCl , 5 EGTA, 10 NaHEPES, 1% branching,andareoflowerorderthantheapicalterminal
3 3 2
biocytin (pH7.4;Sigma).Cellsweresimultaneouslyfilled branches. Thus, whereas the terminal branches spread
withbiocytinduringrecording.Slicesthatcontainedfilled between the superficial parts of layer III (layer IIIa) out
cells were then fixed in 4% paraformaldehyde in 0.1 M intolayerI,theobliquebranchesforthemostpartreside
phosphate-buffered saline (PBS) solution(pH 7.4) at 4(cid:2)C in the middle portion of layer III (layer IIIb). Prior to the
for 2 days. Slices were placed in 0.1% Triton X-100/PBS clustering analyses, manual inspection excluded all
at room temperature for 2 hours, after rinsing with PBS dendrite segments obtained from images of insufficient
(three times, 5–10–15 minutes), and were subsequently quality (Rodriguez et al., 2006, 2008) or with unusually
incubatedwithstreptavidin-Alexa488(1:500;Invitrogen, high or low spine density. Branches were classified
Carlsbad, CA) at 4(cid:2)C for 2 days. After incubation, slices manually as either ‘‘oblique’’ or ‘‘apical terminal’’ through
were mounted onslideswithProlong Goldmountingme- visual inspection. These procedures yielded 152 apical
dium(Invitrogen)andcoverslipped. terminal branches with 23,257 spines and 128 apical
Confocal laser scanning microscopy (CLSM) was per- oblique branches with 19,319 spines, across the seven
formed as described previously (Rocher et al., 2010), neurons. After confocal images had been acquired and
with a voxel size of 0.1 lm2 (cid:3) 0.2 lm. Each neuron’s deconvolved as described above, NeuronStudio and
apicaldendritictreewasimagedinitsentiretyusingmul- Adobe Photoshop CS4 were used to prepare images for
tiple stacks. Raw image stacks then were deconvolved publication.Nootherdigitalmanipulationswereused.
usingAutoDeblur(MediaCybernetics,Bethesda,MD).The
final imageof each neuron(Fig. 1a) was created by inte- Clustering algorithm
gratingalldeconvolvedstackstogetherusingtheVolume Unsupervisedclusteringalgorithmsavoidapriorispeci-
IntegrationandAlignmentSystem(VIAS;Rodriguezetal., fication of either the number of elements per cluster or
2890 TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience
Spineclustering in neurons:morphologicevidence
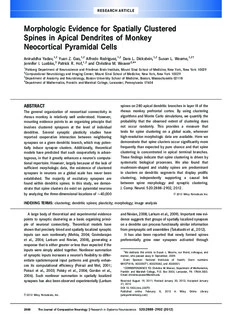
Figure 1. Imaging and automated reconstruction of neurons. a: The xy- and yz-projections of deconvolved CLSM image stacks of a layer
IIIpyramidalneuronfromarea46ofarhesusmonkey,integratedwithVIAS.b:Detailofautomatedreconstructionofdendritictreesusing
NeuronStudio. This region is boxed in a. The box in b denotes the region shown in detail in c,d. c: Dendrite centerline (green line) and
spine detection (red dots) using NeuronStudio, superimposed on the deconvolved data. d: 3-D representation of the dendrite branch and
spinereconstructionshowninc.Scalebars¼50lmina;20lminb;5lminc(appliestoc,d).
the total number of clusters (MacKay, 2003). Iterative In the first iteration, two nearest leaf nodes were
hierarchicalmethodsareoneclassofunsupervised clus- merged to form a higher order node located at the
tering algorithms commonly used in pattern recognition, midpoint of the parent leaf nodes (Sokal and Michener,
data mining, and computational biology (Romesburg, 1958). The merging of nearest nodes repeated in sub-
1984). Hierarchical methods fall into two categories: top sequent iterations, with the parent nodes being either
downandbottomup.Top-downmethodsbeginwithasin- leaf nodes or higher order nodes. Thus, as the
gle cluster containing all of the observations, then split algorithm continued, a node corresponded either to a
the data into smaller clusters if specific conditions are single spine or to a group (cluster) of spines merged
satisfied (Murtagh,1983; Gronau and Moran, 2007). The during previous iterations.
morecommonbottom-upapproachbeginswitheachindi- Figure2billustratesthisiterativecombinationofnodes
vidualdatapoint(‘‘leaf’’),theniterativelycombinesleaves in the form of a dendrogram. The x-axis depicts equally
thatsatisfysomecriteriaofminimumdissimilarityamong spacedleafnodeslabeled1–19,correspondingtothe19
thewholeset. spines on the dendritic branch shown in Figure 2a. The
For our spatially distributed spine data, we chose heightalongthey-axisrepresentsthe1-D‘‘link’’distance
the unweighted pair group method with arithmetic between constituent members of the nodes created by
mean (UPGMA), a simple, iterative, bottom-up hierarchi- eachiteration.
cal clustering method that uses Euclidean distance as Bydefault,UPGMAiteratesuntilallleafnodesarecom-
its minimum dissimilarity criterion. The clustering was binedintoasingleclusterwithhighlydispersedconstitu-
performed separately for each dendrite branch (see ents.Tostoptheclusteringinanobjective,unsupervised
Fig. 2). At the start of the clustering, the 1-D path way, we used a quantitative stopping criterion called the
length of each spine was represented as a leaf node. inconsistency coefficient (IC; Zahn, 1971). The IC
TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience 2891
Yadav et al.
Figure 2. Illustration of UPGMA clustering algorithm. a: Nineteen spines identified automatically along a segment of dendrite branch,
superimposedonthexy-projectionofthedeconvolvedCLSMimage.Eachspineheadisshownasacoloredcircle,withastraightlineillus-
trating its projection onto the dendritic centerline (yellow). Colors signify the final clusters determined by the algorithm shown in f; spines
notbelongingtoaclusterareshowningray.b:DendrogramofthefullUPGMAprocess.Thex-axisdepictsequallyspacedleafnodescor-
respondingtothespinesina;theheightalongthey-axisrepresentsthe1-Ddistancebetweenconstituentmembersofthenodescreated
at each iteration. Colors correspond to the final cluster groupings. Groupings that violate the inconsistency coefficient (see Materials and
Methods), shown in black, were not performed. c: Schematic representation of 1-D spine locations along the dendrite. d: Spine clusters
identified after five iterations of UPGMA (gray horizontal line in b). Colored spines have already been merged into groupings; black spines
havenotyetbeenclassified.e:ClustergroupingsidentifiedattheterminationoftheUPGMAalgorithm,beforepruningunrealisticclusters.
f: Final clustering results, after pruning clusters with the density-based cutoff (see Materials and Methods). Spines identified within each
clusterareshowninasinglecolorandareclassifiedas‘‘clusteredspines’’forsubsequentanalyses.Remainingspines(showningray)are
classifiedassinglets.Scalebar¼2lmina.
measuresthevariabilityinlinksizesofanodeanditscon- As UPGMA progressed, we accepted new nodes only
stituentnodes.TheICfortheithnodeofadendrogramis with an IC less than a prescribed cutoff, thus controlling
givenby the level of dispersal in the extracted clusters. We used
p
ffiffiffi
anICcutoffof0.75,slightlyhigherthan1= 2(cid:5)0:71,the
x (cid:4)x
IC ¼ i k (1) minimumICforanodewithatleastonenonleafconstitu-
i
r
k ent. This cutoff extracted the tightest cluster groupings
where x is the link corresponding to the ith node, and while ensuring that at least one leaf was included in the
i
x and r are the mean and standard deviation of the formation of any node. Moreover, the algorithm was
k k
set of links of all nodes constituting node i from k previ- allowedtogroupleavesintopairsbutcouldonlyaddone
ous iterations, including x itself. For a node consisting leafatatimebeyondthat(Fig.2c–e).
i
of two leaves, with no lower level links against which to UPGMA is an unsupervised algorithm, so we postpro-
compare, the IC is assigned a value of 0. The IC value cesseditsresultstoremoveclustersthatwerenotbiolog-
for a given node is low if the constituent leaf nodes are ically plausible. Clusters having a spine density (spines
grouped together tightly and high if the constituent per micrometer of dendrite) smaller than the branch
nodes are more dispersed. spine density were deemed to be unnphysical. Each
2892 TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience
Spineclustering in neurons:morphologicevidence
unphysicalclusterwasreducediterativelybypruningthe
one spine whose removal provided the largest decrease
in the length of the remaining cluster (Fig. 2f). This pro-
cesscontinueduntilthespinedensityoftheprunedclus-
terwasgreaterthanthebranchspinedensity.
Inouranalyses,aspineclusterisdefinedasagroupof
three or more spines after the completion of UPGMA
analysisandpostprocessing.SpinesbelongingtoUPGMA-
identifiedclusterswerecategorizedas‘‘clusteredspines.’’
Remainingspineswerecategorizedas‘‘singletspines.’’
Monte Carlo simulations and C score
calculation
We used Monte Carlo simulations to quantify the
amountofclusteringobservedineachdendritebranchof
our data relative to the amount expected in equivalent
setsofrandomlyplacedspines.SupposeBwasonesuch
dendritebranchwith1-DlengthLBandnBspines.Wealso Figure 3. Randomizationmethodtoevaluatedegreeofclustering
created M ¼ 5,000 copies of B, randomly placing the n indata.a:FinalUPGMAresultfromFigure2f,asarepresentative
B
spines according to a uniform distribution along the branch to illustrate randomization. Each section of length LB and
n spines was analyzed via UPGMA, resulting in Q spines
lengthofabranchforeachcopy,B,i¼1...M(Fig.3a). B B
i
grouped into clusters. Then, M ¼ 5,000 copies of the branch
UPGMAwasthenperformedontherealbranchBandon
were made, each having length L and randomized locations of
B
each of the randomized copies B. Let Q (cid:6) n be the
i B B the nB spines. Random branch copies are labeled as Bi, with i ¼
numberof clustered spines onthe real branchB, andlet 1,2...M,andthenumberofspinesinclustersforeachrandom
q bethenumberofclusteredspinesonrandomizedcopy copy is labeled q. b: The M q values make up an approximately
i i i
B.Weinterpretedtheset{q}asobservationsofarandom normal distribution. The C-score for each real branch gave the
i i
percentageofthisdistributionthatissmallerthanC.
variable,W ,representingthenumberofclusteredspines T
B
extractedfromeachrandomcopyofbranchB.
The frequency distribution of W over all randomized
B ters occurred more frequently than was expected
copieswasnormalizedtoobtainaprobabilitydistribution,
randomly. If so, then the second goal was to compare
P(W ),ofW .Then
B B quantitatively the characteristics of spine clusters
ZQB (e.g., cluster size, composition of spine types, spine
C ¼ Pðw Þdw ; (2) number per cluster) that were generated by systematic
B B B
biological processes with the characteristics of clusters
0
generatedrandomly.Intheabsenceofmorphologicdata
was the cumulative distribution function of the random
on the precise locations of presynaptic neurons, it is
variable W , with 0 (cid:6) C (cid:6) 1. Hereafter called the ‘‘C-
B B impossibletopredictfunctionalsignificanceonacluster-
score,’’ C gave the proportion of randomized copies of
B by-cluster basis. Instead, we used statistical testing to
B that had fewer spines in clusters than was observed
distinguishindividualdendriticbranchesthatwere‘‘highly
in the real data (Fig. 3b). A C-score close to 1 indicated
clustered,’’ possessing significantly more spine clusters
that the number of clustered spines observed on the
than were expected randomly, from branches that were
true branch B was significantly higher than expected by
‘‘sparsely clustered.’’ This led to our choice of the bino-
chance alone.
mial test (Siegel, 1956), a test that is commonly used
SimilarlytothecalculationofC ,C-scoreswerealsocal-
B when grouping data into two categories. The distribution
culatedforeachrandombranchcopy.Bydefinition,these
of C-scores for real and randomly generated dendritic
C-scores were analogous to P values with P(W ) as the
B branches was divided into two categories by introducing
underlyingdistributionandconsequentlywereguaranteed
a threshold C , 0 (cid:6) C (cid:6) 1. Branches with C-scores
T T
tobeuniformlydistributed(EwensandGrant,2001).
higherthanC wereclassifiedashighlyclustered;allother
T
segments were considered sparsely clustered. A larger
Statistical analysis thresholdC providedamoreconservativeclassificationof
T
The first goal of the statistical analysis was to test a branch as highly clustered. Because C-scores for ran-
whetherthedatasupportthehypothesisthatspineclus- domlygeneratedsegmentswereuniformlydistributed,the
TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience 2893
Yadav et al.
Figure 4. Cluster groupings were largely insensitive to the inconsistency coefficient (IC) cutoff. The x-axis illustrates the 1-D locations of
62 spines along a 50-lm length of dendrite. Each color signifies a group of spines merged into a single cluster by UPGMA, using the IC
criterion cutoffs shown along the y-axis. Cluster groupings were identical for IC cutoffs from 0.75 to 0.95. Above 0.95, smaller clusters
weremergedintolargerones,asdenotedbybracketsandarrows.
proportionsofhighlyclusteredandsparselyclusteredseg- the expected frequency in each cell (Cohen, 1988). From
mentsintherandomizeddatawere1(cid:4)C andC ,respec- theMonteCarlodata,werepeatedthev2test1,000times
T T
tively.Wetestedwhethertheproportionofhighlyclustered with different random samples of 200 data points each.
segments in the real data was significantly higher than Wealsoconductedthev2testoncefortheCscore/spine
1(cid:4)C .Underthenullhypothesisthatrealsegmentswere density pairs from the real data (n ¼ 280), considering
T
distributednodifferentlyfromrandomsegments,theprob- obliqueandtuftbranchestogether.
abilityofanindividualsegmentbeinghighlyclusteredwas Spine cluster statistics were reported as arithmetic
s ¼ 1 (cid:4) C . Then, assuming a binomial distribution, the mean 6 standard error of the mean. Statistical compari-
T
probabilitythatatleastmsegmentsfromatotalofnseg- sons among highly clustered and sparsely clustered
mentswereclusteredwasgivenby branches were performed using the Wilcoxon rank sum
method after analyzing for normality using the Lilliefors
n
P ¼X Csið1(cid:4)SÞðn(cid:4)iÞ; (3) test(Lilliefors,1967).Forallstatisticaltests,nullhypoth-
n i
i¼m eseswererejectedata¼0.05.
where C is the combination operator, equal to the
n i
RESULTS
number of combinations of n objects taken i at a time.
This gave the P value for the binomial test. Seven layer III pyramidal neurons drawn from the dor-
Totest thenull hypothesis thatC-scoreand spine den- solateralPFCofthreeadultrhesusmonkeyswereimaged
sitywereindependent,weconstructedcontingencytables with a voxel size of 0.1 lm2 (cid:3) 0.2 lm, and dendritic
ofC-scorevs.spinedensity.Itisreasonabletohypothesize branches and spines were reconstructed using auto-
that a high spine density would lead to a high C-score; mated software (Fig. 1; see Materials and Methods). We
thereforeourone-sidedalternativehypothesiswasthatC- thenusedourclusteringmethod(Fig.2)andMonteCarlo
score and spine density were positively correlated. For a simulations(Fig.3)toanalyze128apicalobliqueand152
given set of dendrite branches, we first determined the apical terminal branches that included 19,319 and
boundariesofthefourquartilesofC-score(Q1,C,Q2,C,Q3,C, 23,257spines,respectively.
Q )andspinedensity(Q ,Q ,Q , Q ).Then,each As detailed in Materials and Methods, the merging of
4,C 1,D 2,D 3,D 4,D
cell (i,j) of the 4 (cid:3) 4 contingency table was equal to the spine clustersbytheUPGMA algorithm wasgovernedby
numbersofdendritebranchesthatwereinquartileiforC the inconsistency coefficient (IC). We found that cluster
score and quartile j for spine density. Thus, from a set of extraction was largely insensitive to changes in the IC
nbranches,theexpectedfrequencyineachcellwasn/16. cutoff,suggestingthatouralgorithmicimplementationof
A one-tailed v2 test for independence (Ewens and Grant, clusterextractionwasrobust.Figure4illustratesclusters
2001) was used to compute the test statistic (9 df). identified after UPGMA but before the postprocessing.
G*Power 3 (Faul et al., 2007) was used to determine an ExtractedclusterswereunchangedforICcutoffsranging
appropriate sample size (n ¼ 231) so that the test had from 0.75 through 0.95. Above an IC cutoff of 0.95,
95%powertodetectaneffectsizeof0.32,whichwassuffi- smaller clusters were increasingly merged into larger
cient to detect an effect as small as a 2% deviation from ones. Because we wished to be conservative in
2894 TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience
Spineclustering in neurons:morphologicevidence
Figure 5. C-score vs. spine density forrandomized and real data. a: Scatterplot of C-score vs. spine densityfor randomized copies of 32
dendrite branches. Each red circle denotes the clustering result of a branch from the real data, a subset of the data from b. Open black
circles show the C-score for 50 randomized copies of each of these branches. For these randomized data, C-score was independent of
spine density. b: Scatterplot of C-score vs. spine density for all 280 dendrite branches from the real imaged neurons. Red circles denote
apical terminal branches, andblack circles denote oblique branches. The dashed line showsthe best-fit linear regression (r2¼ 0.02),and
thegrayregionrepresentsbranchesthatwereclassifiedas‘‘highlyclustered.’’
identifying spine clusters, we performed our subsequent 0.010). Next, by using simple linear regression analysis,
analyseswithanICcutoffof0.75,justabovetheminimal we attempted to predict C-score from spine density of
p
ffiffiffi
ICof1= 2(cid:5)0:71. these data. Although the slope was significantly greater
than zero (slope ¼ 0.061; t ¼ 2.63; P ¼ 0.0091;
278
The C-score is independent of spine density Y¼0.39þ0.061X),thelowcoefficientofdetermination
for randomly placed spines (r2 ¼ 0.024) indicated that spine density explained very
It is reasonable to hypothesize that high spine density littleoftheobservedvariationinC-score(Fig.5b).
onabranchleadstoahigherC-score.Thatis,aclustering
measure might identify a group of randomly spaced Nonrandom clustering in cortical
spinesashighlyclusteredsimplybecausethespinesare apical dendrites
tightlypacked(highdensity).Wefirsttestedwhetherthis For the remainder of this article, we use the C-score
seemedtobetrueifspineswereplacedrandomlyonthe thresholdC todefinehighlyclustereddendritebranches
T
branch. In particular, the Monte Carlo procedure (see as those exhibiting more clustered spines than expected
Materials and Methods) created 5,000 copies of each randomly (see Materials and Methods). Remaining
real dendrite branch with randomly distributed spines, branches were considered sparsely clustered. Figure 6
1.4 million branches in all. Using 1,000 different random presents typical examples of these branch types. Spines
samples taken from the Monte Carlo population (Materi- classified as lying in clusters are shown in pink; singlet
als and Methods), we tested the null hypothesis that the spinesthatwerenotclassifiedintoclustersareshownin
C-scoreofadendritebranchwithrandomlyplacedspines orange (Fig. 6a,b; Fig. 6c shown in black). A greater pro-
was independent of spine density. In repeating the v2 portionofthespineswasclassifiedaslyinginclusterson
testoneachsample,thenullhypothesisofindependence the highly clustered branch (Fig. 6a,c, C-score 93) than
wasrejectedonly50times.Thisisequaltothefalse-posi- on the sparsely clustered branch (Fig. 6b,c; C-score 44).
tiveerrorratea¼0.05,sotheseresultsstronglysupport The 2-D projections (xy, yz, and xz) may be misleading,
the idea that C-score is independent of spine density which is why it is important that the neurite reconstruc-
when spines are distributed randomly. Figure 5a illus- tion and spine detection algorithms operate in three
trates C-score vs. spine density for a random sample of dimensions. In particular, in 2-D projections, the trav-
thispopulationfrom32dendriticbranches. ersed distance along the dendrite may appear distorted,
Fromthe280branchesoftherealneurons,thenullhy- andspinesmayevenseemtobefalsepositives.Looking
pothesisofindependencewasrejected(v2¼21.71,P¼ atallthreeprojectionshelpstoclarifythedistortionsbut
TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience 2895
Yadav et al.
Figure 6. Examplesoftypicalhighlyclusteredandsparselyclusteredbranches.Shownarexy-,yz-,andxz-projectionsof35-lmsegments
of ahighlyclusteredbranch(a) andasparsely clustered branch(b).Spinesclassified asbelonginginclustersareshowninpink;nonclus-
tered‘‘singlet’’spinesareshowninblack.c:Positionsofspinesonthesebranches,asthe1-Dtraverseddistancealongthedendriticcen-
terline.Clusteredspinesareshowninpink,singletspinesinblack.Scalebars¼5lm.
TABLE1. TABLE2.
Numbersof HighlyClustered Branchesvs. C-Score ClusterStatisticsinApicalTerminalBranches
ClusteringThreshold
Highly Sparsely
Apicalterminal Obliquebranches clustered clustered Pvalue
branches(n¼152) (n¼128)
Spinesin 69.5165.08 53.8268.1221 <0.0001
Highly Highly clusters(%)
clustered clustered Spinespercluster 3.5460.12 3.4360.18 0.006
ThresholdC branches Pvalue branches Pvalue Clusterlength(lm) 0.8060.38 1.1360.50 0.0037
T
Cluster 0.3560.14 0.2060.11 <0.0001
0.9 22 0.049 9 0.90 density(/lm)
0.95 14 0.02 4 0.88 Clusteroccupancy 0.2460.04 0.1860.04 <0.0001
Spinedensity(/lm) 1.81860.74 1.30260.71 0.0021
does not completely eliminate them. Figure 6c clearly
illustrates the spine locations along the length of each attributes of clusters found on highly clustered and
dendrite. sparselyclusteredbranches.
Table 1 summarizes the number of highly clustered
branches as determined by two different values of the Spine cluster statistics
threshold C (0.90 and the more conservative 0.95). We We report spine cluster statistics on the 152 apical
T
testedthenullhypothesisthatthenumberofhighlyclus- terminal branches, for the highly clustered and sparsely
tered branches observed in our data was equal to the clustered groups (Table 2). As expected, the percentage
number that was expected with randomly distributed of spines residing in clusters was greater for highly clus-
spines. For apical terminal branches, the null hypothesis tered branches compared with sparsely clustered
wasrejectedforbothvaluesofC (Pvaluesof0.049and branches (P < 0.006). However, the percentage of
T
0.02 for C ¼ 0.90 and 0.95 respectively). For oblique spines residing in clusters in sparsely clustered
T
branches,the nullhypothesis wasnot rejected for either branches was similar to the percentage calculated from
value of C (P ¼ 0.90 and P ¼ 0.88). Thus our data sup- all 1.4 million simulated branches with randomly spaced
T
port the idea that spines were highly clustered on apical spines (56.6%). The mean number of spines per cluster
terminalbranchesbutnotonobliquebranches.Hereafter was greater on highly clustered branches (P ¼ 0.02),
we use the less conservative clustering threshold C ¼ whereas the mean cluster length was smaller (P ¼
T
0.90 and yet show that significant differences exist in 0.0037). The cluster density (number of clusters per
2896 TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience
Spineclustering in neurons:morphologicevidence
Figure 7. Densityofspineshapesinhighlyandsparselyclusteredbranches.Thefigureshowsthedensityofmushroom-shaped,thin,and
stubby clustered spines per unit branch length (a); singlet spines per unit branch length (b); clustered spines, normalized by the total
length of dendrite over which clusters reside (c); and singlet spines, normalized by the length of dendrite over which singlets reside (d).
Barsshowthemeanandstandarderrorofthemean.Blackandgraybarsrepresenthighlyclustered(n¼22)andsparselyclustered(n¼
130)branches,respectively.*P<0.05,**P<0.01.
micrometer) was also larger on highly clustered Bydefinition,proportionallymorespinesresideinclus-
branches (P < 0.0001). This statistic can be misleading, ters on highly clustered branches than on sparsely clus-
because the number of spines per cluster and cluster tered branches. The larger densities of clustered spines
length varied widely. Thus, we also measured spine of each type on highly clustered branches could be due
cluster occupancy, defined as the ratio of the total to clusters being more numerous in these branches but
length of all clusters on a branch to the branch length. alsocouldbeduetofundamentaldifferencesintherela-
Cluster occupancy was greater on highly clustered tiveproportionsofspinetypeswithinhighlyandsparsely
branches (P < 0.0001). clusteredbranches.Thus,wereexaminedourshapeden-
Figure 7 summarizes our findings on the density of sity results after controlling for the proportion of clus-
the three spine types (mushroom-shaped, stubby, thin) tered spines. After dividing the number of clustered
in apical terminal branches. We report the spine type spines of each type by the total cluster length
density,equaltothenumberofeachspinetypepermicro- (asopposedtothebranchlength),thedensitiesofmush-
meterofbranchlength,forclusteredandsinglet(nonclus- room-shaped and stubby clustered spines were still sig-
tered) spines. The density of clustered spines of all types nificantly greater in highly clustered branches (Fig. 7c),
wassignificantlygreaterinhighlyclusteredbranchesthan by43%and56%respectively.Likewise,asalternativesin-
in sparsely clustered branches (Fig. 7a). On highly clus- gletshapedensities,wedividedthesingletspinecountof
tered branches, the density of mushroom-shaped and eachtypebythetotalbranchlengthminusclusterlength.
stubby clustered spines was nearly twice the density on Still, there was no significant difference in the density of
sparsely clustered branches. In contrast, the density of singlet spines of any shape for highly clustered vs.
mushroom-shaped, stubby, or thin singlet spines did not sparsely clustered branches (Fig. 7d), indicating that the
differ between highly clustered and sparsely clustered increaseinmushroom-shaped,stubby,andthinspineson
branches(Fig.7b). highly clustered branches was concentrated in the
TheJournalofComparativeNeurology|ResearchinSystemsNeuroscience 2897
Description:the U.S. Public Health Service policy on humane care and. Spine clustering in . clustering analyses, manual inspection excluded all dendrite segments