Table Of ContentApplMicrobiolBiotechnol(2017)101:5847–5859
DOI10.1007/s00253-017-8338-x
METHODSANDPROTOCOLS
Evaluation and optimization of PCR primers for selective
and quantitative detection of marine ANME subclusters involved
in sulfate-dependent anaerobic methane oxidation
PeerH.A.Timmers1,2&H.C.AuraWidjaja-Greefkes1&CarolineM.Plugge1,2&
AlfonsJ.M.Stams1,3
Received:22February2017/Revised:7May2017/Accepted:8May2017/Publishedonline:15June2017
#TheAuthor(s)2017.Thisarticleisanopenaccesspublication
Abstract Sincethediscoverythatanaerobicmethanotrophic From these qPCR tests, only certain combinations seemed
archaea (ANME) are involved in the anaerobic oxidation of suitableforselectiveamplification.Afteroptimizationofthese
methane coupled to sulfate reduction in marine sediments, primer sets, we obtained valid primer combinations for the
different primers and probes specifically targeting the 16S selective detection and quantification of ANME-1, ANME-
rRNAgeneofthesearchaeahavebeendeveloped.Microbial 2a/b,andANME-2cinsampleswheredifferentANMEsub-
investigation of the different ANME subtypes (ANME-1; typesandpossiblymethanogenscouldbepresent.Asaresult
ANME-2a,b,andc;andANME-3)wasmainlydoneinsed- of this work, we propose a standard workflow to facilitate
iments where specific subtypes of ANME were highly selectionofsuitableprimersforqPCRexperimentsonnovel
enrichedandmethanogeniccellnumberswerelow.Indiffer- environmentalsamples.
entsedimentswithhigherarchaealdiversityandabundance,it
is important that primers and probes targeting different Keywords Anaerobicoxidationofmethane .AOM .
ANME subtypes are very specific and do not detect other Methanotrophs .ANME .qPCR .Primers
ANMEsubtypesormethanogensthatarealsopresent.Inthis
study, primers and probes that were regularly used in AOM
studiesweretestedinsilicooncoverageandspecificity.Most Introduction
ofthepreviouslydevelopedprimersandprobeswerenotspe-
cificfortheANMEsubtypes,therebynotreflectingtheactual Atmospheric methane (CH ) is the second most important
4
ANMEpopulationincomplexsamples.Selectedprimersthat greenhouse gas on earth and accountsfor 20% of all the in-
showedgoodcoverageandhighspecificityforthesubclades fraredradiationcapturedintheatmosphere(Daleetal.2006).
ANME-1,ANME-2a/b,andANME-2cwerethoroughlyval- Marine sediments produce significant amounts of methane,
idated using quantitative polymerase chain reaction (qPCR). and most methane derives from organic matter degradation
andtoalesserextentfromthermogenicandgeochemicalpro-
cesses (Reeburgh 2007; Thauer and Shima 2008). The pro-
Electronicsupplementarymaterial Theonlineversionofthisarticle
(doi:10.1007/s00253-017-8338-x)containssupplementarymaterial, ducedmethaneonlypartlyreachesthewatercolumnthrough
whichisavailabletoauthorizedusers. seeps,vents,andmudvolcanoesorviadiffusionfromanoxic
sediments and dissolution of methane clathrate hydrates.
* PeerH.A.Timmers Morethan90%oftheannuallyproducedmethaneisoxidized
[email protected] coupledtosulfatereduction(SR)inanoxicmarinesediments
before it reaches the hydrosphere (reviewed in Hinrichs and
1 LaboratoryofMicrobiology,WageningenUniversity,Stippeneng4, Boetius 2002; Knittel and Boetius 2009; Reeburgh 2007).
6708WEWageningen,theNetherlands Anaerobic oxidation of methane (AOM) coupled to SR was
2 CentreofExcellenceforSustainableWaterTechnology,Wetsus, firstdiscoveredinmarinesedimentsatthezonewheregradi-
Oostergoweg9,8911MALeeuwarden,theNetherlands entsofmethaneandsulfateoverlap,thesulfate-methanetran-
3 CentreofBiologicalEngineering,UniversityofMinho,Campusde sition zone (SMTZ) (Martens and Berner 1974; Reeburgh
Gualtar,4710-057Braga,Portugal 1976). Molecular studies showed that most archaeal 16S
5848 ApplMicrobiolBiotechnol(2017)101:5847–5859
rRNA gene sequences that were retrieved from marine systems and microbial mats where the in situ archaeal com-
methane-oxidizing environments belonged to specific clades munitywasinvestigatedusing16SrRNAgeneanalysis.The
in the Euryarchaeota that were named anaerobic majority of these previously developed widely used probes
methanotrophic archaea (ANME) (Hinrichs et al. 1999; and primers such as EelMS932 (Boetius et al. 2000),
Boetius et al. 2000; Orphan et al. 2001). In marine environ- ANME-1-350 (Boetius et al. 2000), ANME2a-647, ANME-
ments,threecladesofANMEwereidentifiedandthesewere 2c-622,andANME-2c-760(Knitteletal.2005)wereindeed
namedANME-1(consistingofsubclustersaandb),ANME-2 suitabletostudyarchaeainvolvedinAOMintheseenviron-
(consisting of subclusters a, b, and c), and ANME-3. The ments.However,itisnotknowniftheseprobesandprimers
ANME-1 cluster is related to Methanomicrobiales and capturethefulldiversitywithinANMEcladesandiftheyare
Methanosarcinales but forms a separate cluster (Hinrichs specificforcertainANMEcladesthatoccurinotherenviron-
et al. 1999), ANME-2 are related to cultivated members of ments.TheANME-3subtypehassofaronlybeenreportedto
the Methanosarcinales (Hinrichs and Boetius 2002), and occurinsomemudvolcanoes(Lösekannetal.2007;Niemann
ANME-3 are most related to Methanococcoides spp. et al. 2006; Omoregie et al. 2008) whereas in most marine
(Knitteletal.2005).ThesubclustersANME-2aandANME- sediments, the ANME subtypes ANME-1, ANME-2a,
2b were subdivided, but they form a coherent clade that is ANME-2b, and ANME-2c and different methanogens are
clearlyseparatedfromANME-2c,andtheyarethereforeoften present and show overlapping regions of occurrence
clusteredasANME-2a/b(Timmersetal.2017)(Fig.1).The (Nunouraetal.2006;Orcuttetal.2005;Orphanetal.2004;
sequencesderivedfromclonelibrariesofthefirststudieswere Pachiadakietal.2011;Roalkvametal.2011,2012;Yanagawa
usedtodevelopprobesforfluorescenceinsituhybridization etal.2011).Therefore,indifferentmarinesedimentsthathar-
(FISH)andprimersforquantitativePCR(qPCR)-basedanal- borahighdiversityofANMEandmethanogens,itisimpor-
ysis.Theseprobesandprimersweremainlyusedtostudyseep tantthatprimersandprobestargetingANMEareveryspecific
a b
Fig.1 aThelowestpercentageof16SrRNAgenesequencesimilarity AOM performing clades (white). Using 1291 sequences from the
between and within ANME clades and the GoM-Arc I clade (that SILVASSUrefNR99database(release119.1)(Quastetal.2013),the
containedANME-2d).Similaritieswerecalculatedusingallsequences tree was constructed with the ARB software package (version arb-
ofthespecificcladesfromtheSILVA16SrRNAdatabaseversionSSU 6.0.1.rev12565) (Ludwig et al. 2004). Trees (bootstrapping value of
r122RefNR(Quastetal.2013)withthedistancematrixmethodofthe 1000trees)werecalculatedwiththeARBneighbor-joiningmethodwith
ARBsoftwarepackagewithsimilaritycorrection(Ludwigetal.2004).b terminalfilteringandtheJukes-Cantorcorrection.Crenarchaeotagroup
Phylogenetictreeoffulllength16SrRNAgenesequencesofarchaeal C3 was used as outgroup. The scale bar represents the percentage of
clades that harbor AOM performing archaea (colored) and other non- changespernucleotideposition
ApplMicrobiolBiotechnol(2017)101:5847–5859 5849
anddonotdetectotherANMEsubtypesormethanogensthat MC3 and Desulfovibrio G11. We also included environ-
arepresent.Itindeedappearedthatpublishedprimerpairsand mental samples from Eckernförde Bay (Baltic Sea,
probeswerelesssuitedforotherenvironments,especiallyin Denmark) which is a gassy diffusive sediment different
quantitative PCR (qPCR) experiments. Thus, new specific from seeps and hydrothermal vents, since methane is pro-
primersemerged,butthedesign,validationandoptimization duced from in situ organic matter degradation (Treude
ofprimersforthedifferentANMEsubcladesisdifficult.This et al. 2005b). This sediment contained ANME-1,
is mainly because the phylogenetic distances are large be- ANME-2a/b, and ANME-2c and methanogens (Timmers
tween ANMEsubclades aswellaswithinANMEsubclades et al. 2015a, b) and is therefore highly suitable to validate
(Knittel and Boetius 2009). With more 16S rRNA gene se- primers and probes on specificity for the different ANME
quences emerging in the database, primers and probes are subtypes. High specificity will enable studies on abun-
continuously developed to detect novel ANME sequences, dance and occurrence of different ANME clades which
orwhenpublishedonesweredeemednotspecific. is important for understanding global methane emissions
In this study, we performed in silico validation of the from marine sediments and other methane-cycling envi-
so far published primers and probes that were used to ronments. The workflow applied for evaluation and opti-
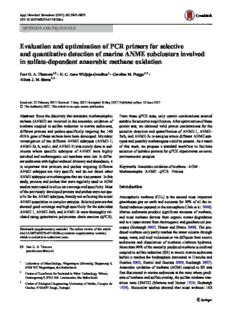
study ANME that performed sulfate-dependent AOM in mization of qPCR primer sets is shown in Fig. 2.
marine sediments. We therefore focussed on oligonucleo-
tides that target the clusters ANME-1, ANME-2a/b (pre-
vious primer sets covering only ANME-2a or only Materialsandmethods
ANME-2b were not tested in this work), and ANME-2c.
For each probe or primer pair, we studied the coverage of InsilicotestingofprobesandprimersReportedprobesand
the target ANME groups, as well as the coverage of non- primersusedinmarineAOMstudiesweretestedforcoverage
target groups. When oligonucleotides seemed suitable, andspecificity,usingtheSILVAProbeMatchandEvaluation
in vitro validation and optimization was done for specific Tool-TestProbe3.0andTestprime1.0services(Klindworth
amplification of ANME-1, ANME-2a/b, and ANME-2c, etal.2013)withtheSILVA16SrRNAdatabaseversionSSU
using quantitative PCR. Validation of primers was done r128 Ref NR (Quast et al. 2013). Only results with 100%
using cloned full-length 16S rRNA gene sequence inserts specificity (0 mismatches) were used for both probes and
ofANME-1,ANME-2a/b,andANME-2carchaea,aswell primers.Primerpairsthatwereamixtureofmultipleforward
as 16S rRNA gene sequence inserts of Methanococcoides orreverse primers weresubmitted witha degeneratebaseto
sp. and genomic DNA from Methanosarcina mazei strain Testprime1.0.Forinstance,primerANME1-395Fconsistsof
Fig.2 FlowchartofqPCR Literature Primer design
approachwhenexistingornewly
designedprimersareusedwitha
complexAOMsample Described Tm Obtain Tm
Gradient qPCR(T )
m
curve analysis
T target = T non target? T target ≠T non target?
m m m m
[DNA] gradient
non-target
[DNA] gradient
target
Sample [DNA] Sample [DNA] R2>0.99% T = T T ≠ T
non- non- msamples mtarget msamples mtarget
target target Efficiency
≥ < ≥90-100%
Control Control
[DNA] [DNA]
amplified amplified
Not OK OK OK Not OK
5850 ApplMicrobiolBiotechnol(2017)101:5847–5859
a mixture of three different primers and ANME1-1417R testedforspecificitywithclonedarchaealinsertsofANME-1,
consistedofamixtureoftwodifferentprimersforincreasing ANME-2a/b, ANME-2c, Methanococcoides sp. (HP-Arch-
coverageofthetargetANME-1group(Miyashitaetal.2009). F02, Genbank ID: HF922275.1), and genomic DNA of
Therefore,wecombinedamaximumoftwoprimersineach M. mazei strain MC3 (DSM-2907) and Desulfovibrio G11
Testprimesubmissionbyreplacingonebasewiththedegen- (DSM-7057),aswellaswithacomplexenvironmentalsample
eratebasethatcoversbothprimers;inthiscasewesubmitted from Eckernförde Bay (EB0). For most primer sets, the first
ANME1-395F(1+2)/ANME1-1417R(1+2)andANME1- strategywastoreproducePCRconditionsasdescribedinthe
395F(3)/ANME1-1417R(1+2)toTestprime(seesequence original literature. When not satisfactory, annealing tempera-
informationoftheprimersinTable1).Thisresultsinadiffer- tureswereoptimizedbyperformingagradientPCRusingall
entcoveragethanwhenallthreeprimersinthisinvitroPCR oftheabovelistedtestsamples.Primersspecificforamplifica-
werecombined.Primerandprobecoverageoftargetandnon- tion of ANME-1, ANME-2a/b, and ANME-2c archaea were
targetgroupsisgiveninTables1and2,respectively. validated. After amplification, specificity was checked by
performingameltingcurveanalysis.Thisconsistedofatem-
Environmental samples and pure cultures Samples were perature gradient (72–95 °C) to obtain the specific melting
taken from Eckernförde Bay (Baltic Sea) at station B (water temperatureofthePCRproducts.PCRproductswithadiffer-
depth28m;position54°31′15N,10°01′28E)duringacruise ent sequence and size will show a different melting tempera-
oftheGermanresearchvesselLittorinainJune2005.Thissam- ture.MeltingcurveanalysisofPCRproductsgivesanaccurate
plingsitehasbeendescribedbyTreudeetal.(2005b).Sediment andsensitivemeasurementoftheamountandthedifferenceof
samplesweretakenwithasmallmulticoresamplerasdescribed thePCRproductsthatwereformedascomparedtothepositive
previously(Barnettetal.1984).Thecoreshadalengthof50cm control. Afterwards, PCR products were also checked for the
andreached30–40cmintothesedimentbed.Immediatelyafter correctsizeona1.5%agarosegel,usingthe1-kbplusladderas
sampling,thecontentofthecoreswasmixedinmultiplelarge sizereference(ThermoScientific).
bottles, which were made anoxic by replacing the headspace
withanoxicartificialseawater.Inthelaboratory,theheadspace
wasreplacedbyCH4(0.15MPa)andbottleswerekeptat4°Cin Results
thedark.M.mazeistrainMC3(DSM-2907)andDesulfovibrio
G11 (DSM-7057) were obtained from the culture collection
In silico testing of probes and primers In silico probe and
(DSMZ,Braunschweig,Germany).
primermatchingwasdonewithpublishedprobesandprimers
to obtain coverage and specificity of target groups (marine
DNA isolation Genomic DNAwas extracted using the Fast
ANME subclades 1, 2a/b, and 2c) and non-target groups,
DNAKitforSoil(MPBiomedicals,Solon,OH)accordingto
allowing zero mismatches (100% specificity). In Table 1, re-
themanufacturer’sprotocolwithtwo45-sbeatbeatingsteps
sultsoftheprimermatching(i.e.,insilicoPCR)areshownfor
using a Fastprep Instrument (MP Biomedicals, Solon, OH).
all primer pairs used in previous studies. Most primer pairs
Afterwards, DNAwas purified and concentrated using the
showedagoodcoverageofthetargetgroupwithlittlecoverage
DNA Clean & Concentrator kit (Zymo Research
of non-target groups. Only primer pair ANME-2aF/ANME-
Corporation, Irvine, CA). The DNA concentrations were ei-
2aR did not have a specific target and primer pair ANMEF/
ther determined with the NanoDrop® ND-2000 (Thermo
907RonlytargetedasmallfractionofANME-3.Theresultsof
FisherScientific,Waltham,MA)ortheQubit2.0fluorometer
probe matching, which does not match primer pairs, but
(ThermoFisherScientific).
matches single oligonucleotide sequences to the SILVA 16S
rRNAgenedatabase,aregiveninTable2.Theseresultsshow
Quantitative real-time PCR PCR amplifications were done
thatasignificantamountofprobesshowzeromismatcheswith
intriplicateinaBioRadCFX96system(Bio-RadLaboratories,
non-target groups, sometimes with a high coverage. Primer
Hercules,CA)inafinalvolumeof25μlusingiTaqUniversal
pairs with highest target group coverage and least non-target
SYBR Green Supermix (Bio-Rad Laboratories), 5μl of tem-
group coverage were tested in vitro using quantitative PCR
plateDNA,and1μlofforwardandreverseprimers(concen-
(qPCR)andaregiveninboldinTable1.
trationof10μM),allaccordingtothemanufacturer’srecom-
mendations.Triplicatestandardcurveswereobtainedwithten-
fold serial dilutions ranging from 2 × 105 (corresponding to Invitrotestingofprimers
1ngμl−1DNA)to2×10−2copiespermicroliterofplasmids
containing16SrRNAarchaealinsertsofANME-1(HP-Arch- ANME-1
D10,GenbankID:HF922261.1),ANME-2a/b(HP-Arch-B12,
Genbank ID: HF922244.1), and ANME-2c (HP-Arch-F07, ANME-1-337F and ANME-1-724R (Girguis et al. 2005)
GenbankID:HF922279.1).Allusedprimerswereextensively showed highest coverage of the target group, with lowest
ApplMicrobiolBiotechnol(2017)101:5847–5859 5851
Reference (Miyashitaetal.2009) (Miyashitaetal.2009) (Girguisetal.2005) (Lloydetal.2011)(Boetiusetal.2000)(Miyashitaetal.2009) (Miyashitaetal.2009) (Vigneronetal.2013) (Miyashitaetal.2009) (Miyashitaetal.2009) (Miyashitaetal.2009) (Vigneronetal.2013) (Girguisetal.2003) (Thomsenetal.2001) on
ze(bp) old. xplanati
si b e
Product 1039 1039 358 219 833 833 - 866 960 960 221 268 816 playedin^hodsfor
%) dis met
( e
ge ar nd
Covera er)3.41.0 er)0.1 er)3.30.9er)1.90.5her)3.3 er)1.90.4her)0.8 er)0.50.10 her)0.3 er)0.2her)2.6 er)1.50.4her)1.0 er)0.60.2her)3.2 er)1.81.60.5her)2.8 er)1.60.425 her)0.20.1-0.30.10.1 inthisstudyBMaterialsa
Coverage(%)Non-target 162.2Methanomicrobia(othEuryarchaeota(other) 12.7Methanomicrobia(oth 168.6Methanomicrobia(othEuryarchaeota(other)140Methanomicrobia(othEuryarchaeota(other)2a/b38.9Methanosarcinales(ot Methanomicrobia(othEuryarchaeota(other)2a/b9.5Methanosarcinales(ot Methanomicrobia(othEuryarchaeota(other)0- 2b42.9Methanosarcinales(ot 2a/b3.2Methanomicrobia(oth2c48.3Methanosarcinales(ot Methanomicrobia(othEuryarchaeota(other)2c18.6Methanosarcinales(ot Methanomicrobia(othEuryarchaeota(other)2c65.6Methanosarcinales(ot Methanomicrobia(othANME-2a/bEuryarchaeota(other)2c60.2Methanosarcinales(ot Methanomicrobia(othEuryarchaeota(other)31.6pMC2A209 Methanosarcinales(otDesulfuromonadalesLokiarchaeotaMBG-D specificity(0mismatches).Primerstested sdescribedbyMiyashitaetal.(2009),see
urethatwereusedforANMEdetection SequenceTarget AACTCTGAGTGCCTCCWA/ANME-CCTCACCTAAAYCCCACT AACTCTGAGTGCCCCCTA/ANME-CCTCACCTAAAYCCCACTAGGTCCTACGGGACGCAT/ANME-GGTCAGACGCCTTCGCTGCTTTCAGGGAATACTGC/ANME-TCGCAGTAATGCCAACACTGTTGGCTGTCCRGATGA/ANME-AGGTGCCCATTGTCCCAA TGTTGGCTGTCCAGATGG/ANME-AGGTGCCCATTGTCCCAA ACGGATACGGGTTGTGAGAG/-CTTGTCTCAGTCCCCGTCTCAGTGCCAGTACTAAGTGC/ANME-TTTCGAGGTAGGTACCCAANME-1411RCGCRCAAGATAGCAAGGG/ANME-CCAAACCTCACTCAGATG AGCACAAGATAGCAAGGG/ANME-CCAAACCTCACTCAGATG TCGTTTACGGCTGGGACTAC/ANME-TCCTCTGGGAAATCTGGTTG (FandRareswitched) CGCACAAGATAGCAAGGG/ANME-CGTCAGACCCGTTCTGGTA GGCUCAGUAACACGUGGA/ANME-CCGTCAATTCCTTTRAGTTT gtheonlineTestprimedatabaseofSILVA,using100% ydesignedprimers(indicatedbynumberinbrackets)a
Table1Primersdescribedintheliterat Primercombination aANME1-395F(1+2)/ANME1-1417R(1+2) aANME1-395F(3)/ANME1-1417R(1+2)ANME-1-337F/ANME-1-724RANME1-628f/ANME1-830raANME2a-426F(1+2)/ANME2a-1242R aANME-2a-426F(3)/ANME2a-1242R ANME-2aF/ANME-2aRANME2b-402F/ANME2b-1251R aANME2c-AR468F(1+2)/ANME2c-AR- aANME2c-AR468F(3)/ANME2c-AR-1411R ANME2c-F/ANME-2c-R Ar-468F/AR736R ANMEF/907R AllprimercombinationsweretestedusinaTheseprimersareamixtureofseparatel
5852 ApplMicrobiolBiotechnol(2017)101:5847–5859
coverage of non-target groups in the in silico analysis product length is optimal between 70 and 200 bp), these
(Table 1). This primer pair was described to be specific for primersseemtobespecificandappropriateforquantification
ANME-1, had strong 3′-mismatches to closely related usingourprotocol(Fig.3),butthelowefficiencymayresult
outgroups, and was previously tested for amplification with inlowsensitivitywhentargetconcentrationsarelow.
Desulfobulbusspp.,Beggiatoaspp.,and28archaealandbac-
terialphylotypescommonlyfoundinseepsediments(Girguis ANME-2a/b
et al. 2005). Here, specificity was tested using qPCR with
genomic DNA of M. mazei strain MC3 and cloned full- For specific detection of the coherent clade ANME-2a/b,
length 16S rRNA gene sequences of ANME-1 and ANME- primer set ANME-2a-426-F and ANME-2a-1242-R
2casDNAtemplate.ThisrevealedthattheANME-1primer (Miyashita et al. 2009) were tested in this work.
pairs were not specific under described reaction conditions. Amplification of the cloned 16S rRNA gene sequence of
TheANME-1primerpairgaveaPCRproductwithgenomic ANME-1 as DNA template only occurred at concentrations
DNAofM.mazeistrainMC3asDNAtemplateconsistingof of>2×10116SrRNAgenecopiesμl-1,andthePCRproduct
two bands, with one having the correct fragment size of showed a different melting temperature at lower template
358bpforthisprimerset.Meltingcurveanalysisshowedthat DNAconcentrations.Onlyathighertemplateconcentrations
(oneofthe)PCRproductsalsohadanidenticalmeltingtem- of ANME-1 cloned sequences of >2 × 102 16S rRNA gene
peraturecomparedtothePCRproductofthepositivecontrol. copiesμl-1,thePCRproductswerevisible(Fig.S5).Cloned
The primer pair also gave multiple PCR products with the ANME-2c 16S rRNA gene sequences as template DNA for
cloned 16S rRNA gene sequence of ANME-2c as template this ANME-2a/b primerpair only showed a PCR product at
DNA, with none of these products having the expected concentrationsof>2×10216SrRNAgenecopiesμl-1asseen
ampliconsizeof358 bp(Fig.S1).Thisalsocounted for the fromthemeltingcurveanalysis,buttheproductquantitywas
cloned16SrRNAgenesequenceofANME-2a/b. toolowforavisibleproductonanagarosegel(Fig.S6).The
Another primer pair was described to be specific for sameresultwasobservedwithclonedMethanococcoidessp.
ANME-1: ANME1-395F and ANME1-1417R) (Miyashita 16S rRNA gene sequences as template DNA. The
et al. 2009). With these designed primers for ANME-1, EckernfördeBaysampleastemplateDNAresultedinaPCR
Miyashita et al. (2009) tested the specificity using genomic product with a melting temperature that corresponded to the
DNA from Methanogenium organophilum and PCRproductoftheclonedANME-2a/b16SrRNAgenese-
Methanomicrobiummobile.DetectionofANMEinmethano- quence as template DNA. Since the environmental sample
genic environments such as methanogenic sludge, rice field EB0 used in this study has a low amount of the ANME-2c
soils, lotus field sediment, and natural gas fields was also subtype(Timmersetal.2015a,b),thisprotocolcanbeapplied
performed(Miyashitaetal.2009).However,underthereport- forthisspecificsample(Fig.3).Althoughthecoverageofthis
edconditionsthatwereappliedtoourEckernfördeBaysam- primersetisnotoptimal(±38%),otherpublishedANME-2a/b
ples,thePCRefficiencywiththeANME-1primerswasonly primer sets were not sufficiently covering the target groups
61.8%andthecalibrationcurveshowedanR2valueofonly (Table1).
0.973.Afteroptimization,mainlychangingannealingtemper-
atures, these values greatly improved (efficiency = 87%, ANME-2c
R2=0.998)andmeltingtemperaturesofPCRproductsfrom
both the cloned 16S rRNA gene sequence of ANME-1 and The primer pair AR468f and AR736r was described to be
from the Eckernförde Bay environmental sample EB0 were specificforANME-2candhasbeentestedforspecificitywith
identical (Fig. S2). For the ANME-1 primer set, genomic Methanosarcinaacetivoransandotherrepresentativearchaeal
DNA from M. mazei strain MC3 and Desulfovibrio G11 as groups commonly found in seep sediments (Girguis et al.
templateDNAdidnotgiveaPCRproductafteroptimization. 2003).Theprimersshowedahighcoverageoftargetgroups
Only when using template concentrations of >2 × 102 16S withlow coverageofnon-target groups(Table1).However,
rRNAgenecopiesμl-1cloned16SrRNAgenesequencesof when we performed qPCR, the primer pair was not specific
ANME-2a/b and ANME-2c as DNA template gave a PCR underdescribedreactionconditions.ItshowedaPCRproduct
product (Figs. S3 and S4). Furthermore, when this cloned withtemplateDNAfromM.mazeistrainMC3,andtheam-
ANME-2c 16S rRNA gene sequence gave a PCR product, plifiedproducthadthesameexpectedproductsizeof268bp
the melting temperature was not the same as for the cloned and the same melting temperature as the positive control
ANME-116SrRNAgenesequenceandsampleEB0astem- (Fig. S7). This was also the case for the cloned ANME-1,
plateDNAandthePCRproduct(s)werenotoftheexpected ANME-2a/b,andANME-2c16SrRNAgenesequence.
sizeof1039bp(Fig.S4).Althoughtheefficiencyoftheprim- TheforwardprimerAR468fwasalsousedinamixtureof
er set was not high, probably due to the length of the PCR threeseparateforwardprimerstoincreasecoverage,together
product (efficiency should be between 90 and 100% and witha newreverse primerANME-2c-AR-1411R(Miyashita
ApplMicrobiolBiotechnol(2017)101:5847–5859 5853
etal.2009).Thisprimerpairindeedshowedhighercoverage subtypes,manywerenotspecificallytargetingthecladesthat
of the target group with low coverage of non-target groups theseprobesandprimersweredesignedfor(Tables1and2).
(Table1).Thisprimerpairhasbeentestedforspecificityusing The non-target 16S rRNA sequences that showed no mis-
genomic DNA from Methanogenium organophilum and matches, especially with the investigated probes, included
Methanomicrobium mobile (Miyashita et al. 2009). some problematic non-targets which are shown in
Detection of ANME in methanogenic environments such as Table 2. These were other marine ANME clades and the
methanogenicsludge,ricefieldsoils,lotusfieldsediment,and GoM-Arc I clade, also known as the AAA archaea (Knittel
naturalgasfieldshasalsobeenperformed,aswasdoneforthe andBoetius2009).Thiscladecontainstherecentlydescribed
ANME-1primers(Miyashitaetal.2009).Inourexperiments, BCandidatusMethanoperedensnitroreducens^thatbelongsto
the primer set showed a PCR product with genomic DNA theANME-2dsubcladethatcoupledAOMtonitrateandiron
fromM.mazeistrainMC3andDesulfovibrioG11aswellas ormanganese reduction (Raghoebarsingetal. 2006;Haroon
with all cloned ANME-1 and ANME-2a/b 16S rRNA gene etal.2013;Ettwigetal.2016;Arshadetal.2015).Thereisnot
sequencesasaDNAtemplate.However,multiplePCRprod- muchknownontheoccurrenceandactivityoftheANME-2d
ucts were obtained, but none had the expected product size subclade and on the overarching GoM-Arc I/AAA clade.
and melting temperatures. This was in contrast with PCR However, the GoM-Arc I/AAA clade has been found to co-
products of the cloned 16S rRNA gene sequence from occur with ANME types that are known to be involved in
ANME-2c as DNA template (Fig. S8). The authors claimed AOM coupled to sulfate reduction, such as ANME-1,
thatitwasindeeddifficulttodesignprimersperfectlyspecific ANME-2a/b, and ANME-2c (Timmers et al. 2016; Mills
forANME-2csequences(Miyashitaetal.2009). et al. 2003; Mills et al. 2005; Lloyd et al. 2006). Therefore,
Primers for ANME-2c were designed by others as well, probes and primers specific for ANME involved in sulfate-
such as ANME-2c-F and ANME-2c-R that showed highest dependentAOMshouldnotmatchwith16S rRNAgenese-
coverageofthetargetgroup(Table1)(Vigneronetal.2013). quencesoftheGoM-ArcI/AAAclade,ifthiscladeco-occurs
Under described PCR conditions, ANME-1, ANME-2a/b, withthe ANME subtypes. Withall ANMEsequences inthe
Methanococcoidessp.,andallnegativecontrolsgaveaPCR SILVA 16S rRNA gene database version SSU r122 Ref NR
productoftheexpectedsize.However,afteroptimization,no (Quastetal.2013),wecalculatedthesimilaritybetweenand
PCRamplificationoftheclonedANME-2a/b16SrRNAgene withinANMEcladesandtheGoM-ArcIcladethatcontained
sequence was observed, although ANME-2a/b was targeted ANME-2d, using the distance matrix method of the ARB
with zero mismatches (Table 1). PCR amplification of the software package with similarity correction (Ludwig et al.
cloned16S rRNAgenesequenceofANME-1asDNAtem- 2004). The lowest similarity was between ANME-1 and
plateonlyoccurredwithtemplateconcentrationsof>2×102 ANME-2ccladesandwas74.2%,whichislowerthanprevi-
16SrRNAgenecopiesμl-1(Fig.S9).ThePCRproductofthe ously reported by Knittel and Boetius (2009) (Fig. 1). The
Eckernförde Bay sample showed a melting temperature cor- lowestsimilaritywithin ANMEcladeswas within ANME-1
respondingtothePCRproductsoftheclonedANME-2c16S andwasonly80.6%(Fig.1).Sincethisinter-andintra-group
rRNA gene sequence. Eckernförde Bay samples have low diversity is high, designing primers that should specifically
copy numbers of the ANME-1 subtype, and therefore, this target ANME subclades without targeting outgroups is
protocol can be used in these types of sediments. Although deemed difficult. New sequences added to the database can
DNA of M. mazei strain MC3 and Desulfovibrio G11 did alsodrasticallychangecoverageandspecificityofpreviously
showaPCRproductwiththeseprimers,themeltingtemper- designedprobesandprimers,andtherefore,probeandprimer
ature did not correspond to the melting temperature of the validationneedstobereconsideredconstantly.Asanalterna-
PCR product of the cloned ANME-2c 16S rRNA gene se- tiveto(orcomplementwith)the16SrRNAgeneasbiomark-
quence,whichisreflectedinthedifferentPCRproductsize. er,onecanusefunctionalmarkergenessuchasthegenefor
Therefore,whenusingtheseprimersforenvironmentalsam- thealphasubunitofthemethylcoenzymemreductase(mcrA)
ples,quantificationofANME-2ccannotbedonewhenmul- thatispresentinallmethanogensandANMEsubtypes.This
tiple PCR products are obtained and when different melting mcrAgeneishighlyconserved,andcomparativephylogenetic
temperatures are obtained that are identical to those of studies have clearly shown that mcrA and 16S rRNA gene-
M. mazei strain MC3. The optimized protocol for the basedphylogenyisconsistent(reviewedinFriedrich(2005)).
ANME-2cspecificprimersisgiveninFig.3. However, it has been recently discovered that not only me-
thanogenicarchaeapossessthemcrAgene(Laso-Pérezetal.
2016).
Discussion Afterinsilicoselectionofonly16SrRNAgenetargeting
primersandprobeswithhighestcoverageoftargetgroupand
Fromall16SrRNAgene-basedpublishedprobesandprimers lowestcoverageofnon-targetgroups,wefoundthatsomeof
that were so far designed to be specific for different ANME theselectedprimersandprobesthatshouldspecificallytarget
5854 ApplMicrobiolBiotechnol(2017)101:5847–5859
9)
0
0
al.2000) al.2000) al.2000) al.2002) al.2000) al.,2005a) dBoetius2 al.2005) al.2005)
Reference (Boetiuset (Boetiuset (Boetiuset (Orphanet (Boetiuset (Treudeet (Knittelan (Knittelet (Knittelet
e
Coverag 2-2.8 1.2-100 0.1 0.2-0.3 0.4-0.7 23.9 0.9 0.8-26.9 0.2-4.3 0.2-0.4 0.1 0.1-0.2 0.6 0.5-1.5 2.4 0.5-5 0.2 3.6 55.7 68.7 0.1-31.6 2.5-8.3 9.1 7.6 2.0 0.4-15.8 5.8 1-1.2 0.9-2.5 0.4-15.8 0.7 2.6 3.7 2.2 2.5
A
Non-target Methanocellales Thermoplasmatales Thaumarchaeota Methanosarcinales(other) Aenigmarchaeota CandidatedivisionYNPFF GOMArcI Thaumarchaeota Crenarchaeota Methanomicrobiaceae Methanosarcinales(other) Thermoplasmata Hadesarchaea ClassWSA2 Methanomicrobia(other) Sulfolobales Crenarchaeota Methanomicrobia(other) GOMArcI Ca.Methanoperedens Methanosarcinales(other) Methanococci MSBL-1 Methanomicrobia(other) Euryarchaeota(other) Methanosarcinales(other) Methanomicrobia(other) Aenigmarchaeota WSA2 Methanosarcinales(other) Euryarchaeota(other) Methanomicrobia(other) Methanosarcinales(other) obia(other)Methanomicr ANME-2a/b
e
g
a
ver 9 6 3 9 3 1 5 3 8 5 3 2
Co 67. 91. 50. 75. 78. 63. 12. 89. 79. 87. 62. 76.
b b b
c a/ b a/ c b a/ c
1 1 1 1 2 2 2 2 2 2 2 2
E- E- E- E- E- E- E- E- E- E- E- E-
et M M M M M M M M M M M M
Targ AN AN AN - AN AN AN AN AN AN AN AN AN
n
o
MEdetecti argetsite 05-322 50-367 32-649 6-880 30-847 –38555 12-729 47-664 22-639
T 3 3 6 8 8 5 7 6 6
N
A
or
f
d C
hatwereuse GGTTCT TGATGC GCTTGG GGGCTT CAACAC GGCCGC GGTCCC AAGCCT TGATTG
cribedintheliteraturet ’’Sequence(5-3) AGCCCGGAGATG AGTTTTCGCGCC TCAGGGAATACT GGCGGGCTTAAC TCGCAGTAATGC GGCTACCACTCG TTCGCCACAGAT TCTTCCGGTCCC CCCTTGGCAGTC
s
e
d
s
e
b
2Pro name E-1-305 E-1-350 E1-632 E-1-862 E1-830 E-2-538 E-2-712 E2a-647 E2c-622
e e M M M M M M M M M
Tabl Prob AN AN AN AN AN AN AN AN AN
ApplMicrobiolBiotechnol(2017)101:5847–5859 5855
6)
1
0
2
al.2005) al.2000) al.2000) al.2000) hleretal.
Reference (Knittelet (Boetiuset (Boetiuset (Boetiuset (Hatzenpic
e
g
Covera 2.7-2.9 0.6-5.3 4.9 2.9 1.2 0.9 1.5 0.7 0.7 4.5 0.1-50 6.9-73 0.2-5.9 0.4 14 12.2 1-56.7 54.8 67.5 0.1 9.1 1-31.6 1-7.2 0.7-9.7 1.1-10 1.3-4.5 1-2.7 0.9 1.2 0.4 0.1 0.1
d
ol
b
Non-target Archaeoglobi Methanosarcinales(other) ANME-2a/b Methanomicrobia(other) Ca.Methanoperedens GOM-ArcI ANME-3 ANME-1b Euryarchaeota(other) Methanosaetaceae Thermoplasmata Methanosarcinaceae(other) Methanomicrobiales(other) Thermococci GOMArcI Ca.Methanoperedens Methanosarcinales(other) GOMArcI Ca.Methanoperedens Methanosaetaceae CandidatedivisionMSBL1 Methanosarcinaceae(other) Methanomicrobia(other) Methanosarcinales(other) Methanococcales Archaeoglobi Methanomicrobia(other) GOMArcI Ca.Methanoperedens Methanosarcinales(other) Methanomicrobia(other) Euryarchaeota(other) stedinthisstudyaregivenin
e
t
s
e
b
age Pro
Cover 86.9 79.5 60.8 27.3 83.3 89.2 61.5 12.5 12.5 6 4.8 1.5 0.8 75 5.7 ches).
at
m
s
mi
c a/b c b c a/b b b c a/b b a/b (0
Target ANME-2 ANME-2 ANME-2 ANME-3 ANME-2 ANME-2 ANME-2 ANME-2 ANME-2 ANME-2 ANME-1 ANME-3 ANME-2 ANME-2 ANME-2 pecificity
s
%
0
0
e 1
argetsit 60-777 40-257 38-556 32-949 A,using
T 7 2 5 9 V
L
SI
of
C e
CC AG CG GT abas
T T C A T at
G A G T C d
TC TG GG TG GC obe
CTT ACC CTC CGT GGGC Testpr
’’(5-3) CCAG TACA ACCA CACC CGTA online
uence CCC CAC GCT CTC TTCT ngthe
Seq CG CC CG AG CG usi
d
2(continued) name E-2c-760 S240 S538 S932 E-2b-729 obeswereteste
Table Probe ANM EelM EelM EelM ANM Allpr
5856 ApplMicrobiolBiotechnol(2017)101:5847–5859
Fig.3 OptimizedqPCR
programsforallarchaealprimer
setsusedinthisstudy
differentANMEcladeswerenotspecificinourinvitroqPCR available, one needs to be sure that the primers and probes
analysis.Mostoftheseprimersandprobescanthereforenot useddonottargetcloserelativesandarespecific.
beappliedtocomplexmicrobialcommunitieswheredifferent Aftervalidationandoptimizationofpublished16SrRNA
ANMEcladesandmethanogensco-occur,whichisthecasein gene targeting primer sets, we found three sets suitable for
mostmarinesediments.Validationwithsequencesfromthein specific and quantitative detection of ANME-1, ANME-2a/
situarchaealcommunityofthenewenvironmentistherefore b, and ANME-2c subclades in a complex marine environ-
mandatory, or when no data on the archaeal community is ment, Eckernförde Bay, where different ANME subtypes
Description:and quantitative detection of marine ANME subclusters involved . 2004). b. Phylogenetic tree of full length 16S rRNA gene sequences of archaeal.