Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs PDF
Preview Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs
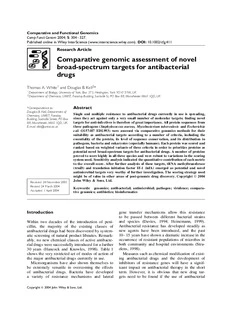
ComparativeandFunctionalGenomics CompFunctGenom2004;5:304–327. PublishedonlineinWileyInterScience(www.interscience.wiley.com).DOI:10.1002/cfg.411 Research Article Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs Thomas A.White1 and Douglas B. Kell2* 1DepartmentofBiology,UniversityofYork,Box373,Heslington,YorkYO105YW,UK 2DepartmentofChemistry,UMIST,FaradayBuilding,SackvilleSt,POBox88,ManchesterM601QD,UK *Correspondenceto: Abstract DouglasB.Kell,Departmentof Single and multiple resistance to antibacterial drugs currently in use is spreading, Chemistry,UMIST,Faraday since they act against only a very small number of molecular targets; finding novel Building,SackvilleStreet,POBox 88,Manchester,M601QD,UK. targetsforanti-infectivesisthereforeofgreatimportance.Allproteinsequencesfrom E-mail:[email protected] threepathogens(Staphylococcusaureus,Mycobacteriumtuberculosis andEscherichia coli O157:H7 EDL993) were assessed via comparative genomics methods for their suitability as antibacterial targets according to a number of criteria, including the essentiality of the protein, its level of sequence conservation, and its distribution in pathogens,bacteriaandeukaryotes(especiallyhumans).Eachproteinwasscoredand ranked basedonweightedvariantsofthesecriteriainordertoprioritizeproteins as potentialnovelbroad-spectrumtargetsforantibacterialdrugs.Anumberofproteins provedtoscorehighlyinallthreespeciesandwererobusttovariationsinthescoring systemused.Sensitivityanalysisindicatedthequantitativecontributionofeachmetric to the overall score. After further analysis of these targets, tRNA methyltransferase (trmD) and translation initiation factor IF-1 (infA) emerged as potential and novel antimicrobialtargetsveryworthyoffurther investigation.Thescoringstrategyused might be of value in other areas of post-genomic drug discovery. Copyright 2004 John Wiley & Sons, Ltd. Received:24November2003 Revised:24March2004 Keywords: genomics; antibacterial; antimicrobial; pathogen; virulence; compara- Accepted:1April2004 tive genomics; antibiotics; bioinformatics Introduction gene transfer mechanisms allow this resistance to be passed between different bacterial strains Within two decades of the introduction of peni- and species (Davies, 1994; Heinemann, 1999). cillin, the majority of the existing classes of Antibacterial resistance has developed steadily as antibacterial drugs had been discovered by system- new agents have been introduced, and the past aticscreening ofnatural product libraries. Remark- 10–15 yearshaveshownadramaticincreaseinthe ably, no new chemical classes of active antibacte- occurrence of resistant populations of microbes in rialdrugsweresuccessfullyintroducedforafurther both community and hospital environments (Stru- 30 years (Hancock and Knowles, 1998). Table 1 elens, 1998). shows the very restricted set of modes of action of Measuressuchaschemicalmodificationofexist- the major antibacterial drugs currently in use. ing antibacterial drugs and the development of Microorganisms have also shown themselves to inhibitors of resistance genes will have a signif- be extremely versatile in overcoming the effects icant impact on antibacterial therapy in the short of antibacterial drugs. Bacteria have developed term. However, it is obvious that new drug tar- a variety of resistance mechanisms and lateral gets need to be found if the use of antibacterial Copyright2004JohnWiley&Sons,Ltd. Assessmentofnovelantibacterialtargets 305 Table1.Modeofactionoftheprincipalestablishedantibacterialdrugs Drug/class Functioninhibited Moleculartarget β-Lactams Peptidoglycansynthesis Transpeptidasesandcarboxpeptidases Bacitracin Peptidoglycansynthesis Undecaprenylpyrophosphate D-Cycloserine Peptidoglycansynthesis D-alanineracemaseandD-alanyl-D-alaninesynthetase Fosfomycin Peptidoglycansynthesis UDP-N-acetylglucosamineenolpyruvyltransferase Glycopeptides Peptidoglycansynthesis Cellwallpeptidoglycan Quinolones DNAreplication/transcription GyraseandtopoisomeraseIV Rifamycins Transcription RNApolymerase Aminoglycosides Proteinsynthesis 30Sribosomalsubunit Chloramphenicol Proteinsynthesis 50Sribosomalsubunit Fusidicadid Proteinsynthesis ElongationfactorG Macrolides Proteinsynthesis 50Sribosomalsubunit Oxazolidinones Proteinsynthesis 50Sribosomalsubunit Streptogramins Proteinsynthesis 50Sribosomalsubunit Tetracyclines Proteinsynthesis 30Sribosomalsubunit Mupirocin ChargingofisoleucyltRNA IsoleucyltRNAsynthetase Sulphonamides Folatesynthesis Dihydropteroatesynthetase Trimethoprim Folatesynthesis Dihydrofolatereductase AfterChopraetal.(2002). drugs is to continue successfully (Schmid, 1998). It has been shown that such data-driven strate- To this end, genomic approaches are providing a gies can be used to identify novel drug targets new strategy by revealing new molecular targets (Spaltmann et al., 1999). A number of metrics are that are giving rise to novel antibacterial agents chosen which should be properties of a potential (Allsop and Illingworth, 2002; Dougherty et al., drug target, such as essentiality and specificity. 2002; Haney et al., 2002; Isaacson, 2002; Ji, 2002; Each potential target in a genome of interest is McDevitt and Rosenberg, 2001), as these new scored for these properties. These scores can be agents are unlikely to face the current problems weighted differently to add more or less emphasis to any particular property. This scoring system can of established mechanisms of resistance (McDevitt be tuned so that targets which have already been and Rosenberg, 2001). In anti-infective research, identified score highly, showing that the scoring the inevitable selection for resistant strains means system iscapableofidentifyinguseful targets.Pre- that drugs with multiple targets may be preferred viously unidentified genes may also score highly, (e.g. multiple penicillin-binding proteins or multi- andthesecanbeprioritizedaspotentialdrugtargets ple forms of two-component systems; Stephenson forfurtherstudy.Thetop-scoring geneinthestudy and Hoch, 2002; Stephenson and Hoch, 2004). In carried out by Spaltmann et al. (1999) on antifun- other pharmaceutical areas it is encouraging that gal targets was α,α-trehalose-phosphate synthase, the rational utility of traditional targets is being a gene which had never before been suggested confirmed by systematic knock-out studies (Zam- as a potential drug target. This shows that post- browicz and Sands, 2003). genomic research has much to offer in terms of Withthereleaseofdatafromnumeroussequenc- novel target identification (Allsop and Illingworth, ing projects, the number of potential drug targets 2002; Buysse, 2001; Dougherty et al., 2002; Glass has increased massively. However, not all of these et al., 2002; Haney et al., 2002; Isaacson, 2002; molecules will become drug targets (Hopkins and Ji, 2002; Knowles and King, 1998; McDevitt and Groom, 2002), and the big challenge is to select Rosenberg,2001;Payneet al.,2001a,2001b;Will- the targets most relevant for a given situation (Ter- ins et al., 2002). stappen and Reggiani, 2001). In the present study a number of criteria were Machine learning methods seek to devise new chosenonwhichtocharacterizeproteinsastargets. ideas and hypotheses from more or less unstruc- These were suggested by the extensive literature tured data (Gillies, 1996; Kell and Oliver, 2004; on the subject (see e.g. Alksne, 2002; Allsop and Mitchell, 1997; Mjolsness and DeCoste, 2001). Illingworth, 2002; McDevitt and Rosenberg, 2001; Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. 306 T.A.WhiteandD.B.Kell Projan, 2002; Spaltmann et al., 1999; Terstappen hit against a look-up table that listed the classifi- and Reggiani, 2001). A full list of the criteria used cation of the organism (http://ca.expasy.org/cgi- is given in the Methods section. bin/speclist). A list of bacteria treated as pathoge- nic in this study is given in Table 2. Bacteria may or may not act as pathogens, depending on the cir- Methods cumstances and the host, and so the list given here covers a broad range of pathogens but is perhaps not completely comprehensive. Data collection and motives The presence of homologues in mice was con- Datawerecollectedfromthreepathogenicbacterial sideredimportantnotonlyasthiswillallowtargets species, Staphylococcus aureus, Escherichia coli whichare present inhigher organisms tobefurther O157:H7 EDL993 and Mycobacterium tuberculo- down-weighted, but also because further down the sis. These species were chosen as they represent line the target’s absence in mice will make animal a broad cross-section of bacterial types. Targets trials more effective. Lactobacillus spp. are con- which prove to score well in these three species sidered to beneficial or probiotic bacteria, so using will probably be good targets across a broad spec- thismetricmightbeabletoprioritizetargetswhich trum of pathogens. diminish any unwanted side-effects of a new drug. The entire set of sequences of proteins encoded The scores of BLAST hits against pathogens by S. aureus, E. coli O157:H7 EDL993 and M. were also parsed to find how well conserved tuberculosis were downloaded from the NCBI a particular gene is amongst pathogens. Obvi- website (http://www.ncbi.nlm.nih.gov/PMGifs/ ously a protein that is well-conserved across many Genomes/micr.html). Each protein was then char- pathogens will make a better target for broad- acterized by a number of criteria which could then spectrum antibacterial drugs. A high degree of be used to prioritize the most suitable proteins as conservation may also mean that mutations in the potential antibacterial targets. protein are not tolerated, such that resistance is A Perl program carried out most of the char- less likely to emerge. The numbers of identical acterization automatically (see Figure 1 for an residues in each pathogenic hit compared to the overview).Eachproteinwasparsedtofindthegene query sequence were summed and then divided by index (gi) number and name of the protein. If the the number of hits against pathogens. This number function of the protein was known, or if a function was normalized by dividing by the length of the had been assigned to the protein on the basis of query sequence, to give a ratio of conservation for sequence homology, then this was noted. this protein across pathogens. Each protein was then submitted to a BLAST The query protein was submitted to BLAST (Altschul et al., 1990, 1997) search (BLASTp, separately against the human genome (protein usingdefault parameters except foran‘expectation sequences) (ftp://ftp.ncbi.nih.gov/genomes/H value’of0.01)againstalocalcopyoftheSwissProt sapiens/protein/) and the number of hits was database (ftp://ftp.ebi.ac.uk/pub/). The SwissProt recorded. The closest hit against a human protein database was used because it is well curated, was also recorded, with a ratio of similarity given well annotated, non-redundant, and since entries bythenumberofpositiveresiduematches(matches are easily parseable due to its consistent format. where amino acids are identical or have similar There also exist a large number of associated files biochemical properties) divided by the length of and websites which use SwissProt-style codes (for the query sequence. The number of positives was species and gene/protein names). Using SwissProt chosen so as to err on the side of caution. Any therefore allows these resources to be integrated drug designed against a particular bacterial protein easily into the program, thus making efficient may act just as well against a human protein, even automation possible. if certain key residues are not identical. Similar- The results of each BLAST search were parsed ity of residues may be enough for activity. This to find how many homologues of this protein metric was included so that potential targets which existed in bacteria, pathogenic bacteria, eukary- were not so similar to human proteins would not otes, mice and Lactobacillus. This was done by be so heavily penalized. Even if a human homo- comparing the SwissProt species ID code of each logue does exist,itmaystillbepossible (e.g. using Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. Assessmentofnovelantibacterialtargets 307 Read in genome from For each query protein file and split into carry out a restricted individual protein BLAST search against sequences (FASTA essential genes in Bacilus format) subtilis, Escherichia coli K12, Mycobacterium tuberculosis and Staphylococcus aureus (expectation value 0.01) Take each protein sequence in turn and parse to find the gene name, gi number and For each query protein whether a function has carry out a restricted been assigned to the BLAST search against protein virulence genes in Bacillus anthracis, Escherichia coli O157:H7 EDL993, Mycobacterium tuberculosis, Neisseria BLAST each protein Meningitidis and against the SwissProt Staphylococcus aureus database (expectation (expectation value 0.01) value 0.01) For each query protein Parse the BLAST carry out another BLAST output from each search of SwissProt (with protein to find higher expectation value taxonomic distribution 1×10-10) and parse output of protein and its to find any hits which are conservation in known antibacterial targets pathogens or which have a structure in the Protein Data Bank BLAST each protein against all protein sequences from Output the information for human (expectation each protein to file. This value 0.01). Find can then be used to score number of human and rank proteins homologues and according to potential as similarity of closest novel broad-spectrum human hit. drug targets Figure1.Flowchartillustratingtheprocessofdatacollection structure–activity relationship studies) to design a as a target or structural similarity it was thought drug which targets only the bacterial version of the safer to report only very close homologues. protein. After running the BLAST algorithm, the output The query gene was then again submitted to was parsed to find whether the query gene was the BLAST program to find homologues which homologous to a known antibacterial target. This are known antibacterial targets or whose structures was done by comparing the SwissProt gene ID have been deciphered. This time an ‘expectation against a list of SwissProt IDs (from the ExPASy value’of1×10−10 wasused,astoinfersuitability website:http://ca.expasy.org/enzyme/)ofproteins Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. 308 T.A.WhiteandD.B.Kell Table2.Listofbacteriatreatedaspathogenicinthisstudy Acinetobactercalcoaceticus Klebsiellapneumoniae Shigelladysenteriae Bacillusanthracis Legionellapneumophila Shigellaflexneri Bacilluscereus Leptospirainterrogans Staphylococcusaureus Bordetellapertussis Listeriamonocytogenes StaphylococcusaureusstrainMu50/ATCC700699 Borreliaburgdorferi Moraxellacatarrhalis StaphylococcusaureusstrainMW2 Brucellaabortus Moraxellalacunata StaphylococcusaureusstrainN315 Brucellamelitensis Mycobacteriumleprae Staphylococcuscapitis Brucellasuis Mycobacteriumtuberculosis Staphylococcusepidermidis Campylobacterjejuni Mycoplasmafermentans Staphylococcussaprophyticus Chlamydiamuridarum Mycoplasmagenitalium Streptococcusagalactiae Chlamydiapneumoniae Mycoplasmahominis StreptococcusagalactiaeserotypeIII Chlamydiatrachomatis Mycoplasmapenetrans StreptococcusagalactiaeserotypeV Clostridiumbotulinum Mycoplasmapneumoniae Streptococcusmutans Clostridiumperfringens Neisseriagonorrhoeae Streptococcuspneumoniae Clostridiumtetani Neisseriameningitidis Streptococcuspyogenes Corynebacteriumdiphtheriae NeisseriameningitidisserogroupA StreptococcuspyogenesserotypeM18 Enterococcusfaecalis NeisseriameningitidisserogroupB StreptococcuspyogenesserotypeM3 Enterococcusfaecium NeisseriameningitidisserogroupC StreptococcuspyogenesserotypeM5 EscherichiacoliO111:H− Pasteurellamultocida Treponemapallidum EscherichiacoliO127:H6 Propionibacteriumacnes Tropherymawhipplei EscherichiacoliO157:H7 Proteusmirabilis Ureaplasmaurealyticum EscherichiacoliO6 Providenciarettgeri Vibriocholerae Flavobacteriummeningosepticum Providenciastuartii Vibrioparahaemolyticus Francisellatularensis Pseudomonasaeruginosa Vibriovulnificus Fusobacteriumnucleatum Rickettsiaconorii Wolinellarecta Haemophilusducreyi Rickettsiaprowazekii Wolinellasuccinogenes Haemophilusinfluenzae Salmonellacholerae-suis Xanthomonasmaltophilia Haemophilusparainfluenzae Salmonellaenteritidis Yersiniapestis Helicobacterpylori Salmonellatyphi HelicobacterpyloriJ99 Salmonellatyphimurium that are known antibacterial targets (Chittum and homology to a protein of known structure is likely Champney, 1995; Egebjerg et al., 1989; Kornder, to have a similar structure (although this is not 2002; Lin et al., 1997; Neu and Gootz, 1996; always true) and so may be favoured as a potential Schnappinger and Hillen, 1996). Of course, not all novel drug target. current drug targets are perfect examples; indeed, Each protein was then submitted to several many of the drugs that target them are toxic to more restricted BLAST searches against selected humans and resistance has begun to emerge in bacterial genomes. The BLAST searches were many cases. Nevertheless, treatments which utilize restrictedbyginumber;specificallytheginumbers these targets have been shown to be effective in of genes found to be essential or involved in disease control, and so novel targets possessing virulence. These genomes chosen are listed in similar characteristics to known targets may be Table 3. useful. These genomes were selected as they cover a The SwissProt species and protein ID codes of wide range of bacterial types, and also because each hit in the BLAST results were compared theyarewellcharacterizedandareamongstthefew to a look-up table (ftp://beta.rcsb.org/pub/pdb/ species for which this work has been carried out to uniformity/derived data/) tofind out whetherany any great extent. For those species for which this homologues of the query gene had an entry in the kindofworkhasnotbeendone,genomicsmethods PDB database (ftp://ftp.ncbi.nih.gov/genomes/H may allow us to predict essentiality or involve- sapiens/protein/). A protein with a known struc- mentinvirulence.Proteinsthathavesignificanthits tureismoreattractivefromthepointofviewoffur- against essential genes or genes involved in viru- therresearch,asstructure-baseddrugdesigncanbe lence are likely to have the same characteristics carried out straightaway. A protein with sequence themselves and so may score highly as potential Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. Assessmentofnovelantibacterialtargets 309 Table 3. List of genomes used for restricted BLAST the importance, or so-called control coefficients searches against essential genes or genes involved in (http://dbk.ch.umist.ac.uk/mca home.htm; Fell, virulence 1996; Heinrich and Schuster, 1996; Kell and West- erhoff, 1986), by which each enzyme controls the Essentialgenes Genomes Bacillussubtilis(Kobayashietal.,2003) fluxthroughametabolicpathway,butcaninfactbe EscherichiacoliK12 used tofind the relative importance of any variable (http://www.shigen.nig.ac.jp/ecoli/pec/ which contributes to a total. The equation giving About.html) thesensitivityofoverallmetricAtoindividualmet- Mycobacteriumtuberculosis(Sassettietal., ric v is given by (equation 1) 2003) i Staphylococcusaureus(Forsythetal.,2002) Virulencegenes ∂A v ∂lnA Genomes Bacillusanthracis(HoffmasterandKoehler, CiA = ∂v · Ai = ∂lnv (1) 1999;Koehler,2002) i i EscherichiacoliO157:H7EDL993(Brunder Here a more discretized sensitivity analysis was etal.,2001;SharmaandDean-Nystrom, 2003;Stuberetal.,2003;Wangetal.,2002) done for each target by taking the score of each Mycobacteriumtuberculosis(Triccasand metricofthetarget,finding1%ofthisscore,divid- Gicquel,2000) ing this number by the total score and multiply- Neisseriameningitidis(Sunetal.,2000) ing by 100. When this is done for all metrics, Staphylococcusaureus(Dunmanetal.,2001) these ‘contributions’ sum to 1. Thus, sensitivity analysis asks, ‘By altering the score of one vari- able by 1%, what percentage change would this drug targets. The more ‘model’ genomes in which induce in the total score?’. These sensitivity analy- the gene is found to be essential, the more likely ses could clearly show when some variables were it is that this gene is indeed essential for the query exerting too much or too little influence on the species, and also has greater potential as a target total score and therefore the weights could be opti- for a broad-spectrum antibacterial drug. mized accordingly. This novel approach proved Having assigned each gene in the query genome very useful in carefully modifying the scoring values for a number of characteristics, these values systems. could then be weighted, summed and ranked to Using different weighting schemes also allowed produce a list of high-priority potential targets. theanalysisofhowrobustaparticularhigh-ranking This ranking approach was used instead of a target was to the weighting scheme. Clearly, a machine learning-based approach, as the ‘training targetwhichscoreshighlyduetohavingfavourable set’ofknownantibacterialtargetsisverysmalland characteristicsinonehighlyweightedmetricisless not necessarily optimal (see Introduction). While good than one which ranks highly under a number the ranking approach is more subjective, it does of different scoring systems. allow targets to be prioritized which score better For each of five different scoring systems according to our metrics than currently known (Table 4) used on S. aureus, E. coli O157:H7 targets. EDL993 and M. tuberculosis the top 20 ranking targets were recorded. These top 20 lists could Assigning weights and the robustness of target then be checked against each other to see whether prioritization robust targets had emerged. The top 20 lists were A number of different weighting schemes were thencross-checked toseewhether anytargets were tried so that theweighting scheme could be refined robust in all three species (see Table 5). This ‘vot- toreflecttherelativeimportanceofthevariousmet- ing’methodapproachcanbeseenascombiningthe rics. After a weighting scheme was run on the raw output of several weak learners, which is known to data, the scores for each metric could be summed be a very effective approach to data mining (Bauer and the total scores of the targets then ranked. and Kohavi, 1999; Dietterich, 2000; Hastie et al., The refinement of the weightings was done by 2001). carrying out a sensitivity analysis on the metric The first scoring system was designed to give scores for the top few ranking targets. Sensitivity most influence to those metrics which were felt to analysis is more normally used in biology to find be the most important and least influence to those Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. 310 T.A.WhiteandD.B.Kell n o 1 1 1 ati00 1 1 1 1 1 1 1 1111 11111 1 5 as as as erv×4 as as as as as as as asasasas asasasasas as me me me nso me me me me me me me memememe mememememe me Sa Sa Sa Corati Sa Sa Sa Sa Sa Sa Sa SaSaSaSa SaSaSaSaSa Sa oooo nnnn ifififif 0000 4 as1 as1 as1 as1 as1 as1 as1 as1 as1 as1 as1 yes,yes,yes,yes, as1as1as1as1as1 as1 me me me me me me me me me me me 0if0if0if0if mememememe me a a a a a a a a a a a 5500 aaaaa a S S S S S S S S S S S 1111 SSSSS S oooo onnnn ifn0if0if0if0if 3 as1 as1 as1 as1 as1 as1 as1 as1 as1 as1 as1 as1as1as1as1 es,0yes,yes,yes,yes, as1 ts m me me me me me me me me me me me memememe ify0if0if0if0if me ge e Sa Sa Sa Sa Sa Sa Sa Sa Sa Sa Sa SaSaSaSa 5010101010 Sa r t a s t y gsystemontherankingof Scorings 2 100/No.ofcopiesofgene (100/No.ofhomologuesin×bacteria)No.ofhomologuesinpathogens ×[(100/89)No.ofdistinctpathogenswith−homologues](No.ofdistinctbacteriawith−homologuesNo.ofdistinctpathogens) ×Conservationratio100 100/(No.ofeukaryotichomologues+1) 100/(No.ofhumanhomologues+1) −100(proximityratio×100) −100(No.ofmousehomologues+1) 100ifabsent,0ifpresent 100ifyes,0ifno 100ifyes,0ifno 100ifyes,0ifno100ifyes,0ifno100ifyes,0ifno100ifyes,0ifno 100ifyes,0ifno100ifyes,0ifno100ifyes,0ifno100ifyes,0ifno100ifyes,0ifno 100ifyes,0ifno n ori ct sttheinfluenceofthesc 1 20/No.ofcopiesofgene (50/No.ofhomologuesin×bacteria)No.ofhomologuesinpathogens ×[(500/89)No.ofdistinctpathogenswith−homologues](No.ofdistinctbacteriawith−homologuesNo.ofdistinpathogens) ×Conservationratio200 50/(No.ofeukaryotichomologues+1) 50/(No.ofhumanhomologues+1) −100(proximityratio×100) −10(No.ofmousehomologues+1) 10ifabsent,0ifpresent 50ifyes,0ifno 20ifyes,0ifno 50ifyes,0ifno50ifyes,0ifno20ifyes,0ifno20ifyes,0ifno 10ifyes,0ifno20ifyes,0ifno20ifyes,0ifno20ifyes,0ifno20ifyes,0ifno 10ifyes,0ifno e t coringsystemsusedto BacillussubtilisEscherichiacoliK12MycobacteriumtuberculosisStaphylococcusaureus BacillusanthracisEscherichiacoliK12MycobacteriumtuberculosisNeisseriameningitidisStaphylococcusaureus s t en et Table4.Thefivediffer Metric Copynumber Distributionofhomologues Speciesdistribution Conservationinpathogens Eukaryotichomologues Humanhomologues Proximitytoclosesthumanhomologue Mousehomologues Lactobacillushomologues Functionknown Homologoustoknowntarg Homologoustoessentialgenein: Homologoustovirulencegenein: HomologoustoPDBentry Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. Assessmentofnovelantibacterialtargets 311 Table5.Theoveralltoptenrankingtargets Rank Genename/description Robustness Totalscore 1 tRNAmethyltransferase(trmD) 15 13391 2 UDP-N-Acetylmuramate-L-alanineligase(murC) 15 13229 3 UDP-N-acetylglucosamine1-carboxyvinyltransferase(murA)∗ 13 13059 4 TranslationinitiationfactorIF-1(infA) 14 13019 5 DNApolymeraseIII,αchain(dnaE) 13 12992 6 30SribosomalproteinS4(rpsD)∗ 11 12779 7 UDP-N-acetylmuramoylalanine-D-glutamateligase(murD) 11 12766 8 50SribosomalproteinL10(rplJ) 11 12755 9 Chromosomalreplicationinitiatorprotein(dnaA) 10 12716 10 UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diaminopimelateligase(murE) 9 12573 Thesetargetsrankhighlyinallthreespeciesusedandrankinthetop20sofmostofthescoringsystemsused.Robustnessishowmanytimes thegeneranksinthetop20underfivedifferentscoringsystemsacrossthethreespeciesused,givingamaximumrobustnessscoreof15. Totalscoreisthesumofthescoresforthistargetinallscoringsystemsacrossallspeciesused.Themaximumpossibletotalscoreis24120. ∗Indicatesthatthegeneisaknowntargetofanantibacterialdrug(murAistargetedbyfosfomycinandrpsDisatargetoftetracyclines). felt to be least important. Homology to essential searchedtofindanyconservedmotifsnotidentified genesinM.tuberculosis andS. aureus,andhomol- by BLAST, which could be used to find more ogy to virulence genes in Bacillus anthracis were distantly related homologues of the query gene. weightedlowerthanhomologytoessentialandvir- This approach was able to identify any human ulence genes in other organisms. This was done to sequences which, although not closely related in reflect the quality of the data for these organisms, terms of sequence homology, could be very sim- asdifferentmethodswereusedandlistsofessential ilar in terms of structure and biochemical proper- and virulence genes are not always complete. ties to the query gene. Multiple sequence align- Underthesecondscoringsystemallmetricswere ments and phylogenetic trees were created using weightedequally,sothatamaximumscoreforone ClustalX (Thompson et al., 1997) and Mega2.1 metric would be the same as for another. For the (Kumar et al., 2001). This was done to deter- other three scoring systems most of the metrics mine how distinct the genes in these pathogens were weighted as under the first system. However, werefrom thosehomologues innon-pathogens and in the third scoring system homology to virulence eukaryotes, and just how well the ‘active sites’ genes was given greater influence, in the fourth of these genes were conserved across the differ- homology to essential genes was given greater entpathogenicspecies.Theavailableliteraturewas weight, and in the fifth the level of conservation of also searched to gain more insights into these sug- thetargetinpathogenswasgivenmoreimportance. gested targets. Further investigation of high-scoring targets Results and discussion Having narrowed down the number of potential Scores drug targets using the methods outlined above, the highest-scoring targets could then be investi- According to the scoring systems used, the major- gated in greater detail. The top genes were again ityofgenesinS. aureus,E. coli O157:H7EDL993 subjected to a BLAST search against the Swis- andM.tuberculosis wouldmakeverypoorantibac- sProt database to determine in which pathogens terialtargets(seeFigures 2–4).Inallthreebacteria they were present. The databases Genbank, EMBL there are also only a few high-scoring genes. The and DDBJ were also searched via ENTREZ highest ranking of these seem to be fairly robust (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) and tend to rank in the top 20, regardless of which to find whether or not a copy of the query gene scoring system is used (see Table 5). existedinaspecificpathogen,incasethishadbeen It is also apparent there are no targets which are missed by searching only the SwissProt database. perfect in every way. To obtain a perfect score PROSITE (http://us.expasy.org/prosite/) wasalso in the present metrics a target should be present Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. 312 T.A.WhiteandD.B.Kell in one copy, be present in all pathogens but not terms of the properties an antibacterial drug tar- in non-pathogens, eukaryotes or humans. It should get should possess. Hence, the first scoring sys- be perfectly conserved across all pathogens. Its tem rates as very important the properties ‘species functionshouldbeknown,itshouldbehomologous distribution’, ‘conservation in pathogens’, ‘similar- to a known target, homologous to essential and ity to human’ and ‘homology to essential genes’. virulence genes in all the model genomes used A target which does not perform well on any one and its structure should be known. Here even the of these criteria will probably not make a good highest-ranking targets achieve only some 50% of drug target. The emphasis accorded to these prop- the perfect score. erties means that targets which are not present in This is perhaps discouraging, as it means that a wide range of pathogens, are not well conserved, there is little possibility for the development of are very similar to targets in humans, or are not a ‘magic bullet’ drug that is highly effective, essential will not be able to score highly and thus specifically targets only pathogens, is easy to will not be prioritized. The metric ‘species distri- develop and is immune to the problems of emerg- bution’ is weighted so that a target will receive the ing resistance. However, this never was a likely maximum score if it is present in all the bacteria prospect. treated as pathogenic by this study and in no non- The unusual peaks in the distribution graphs pathogens. It is unlikely that this maximum would are due to genes of unknown function that, when ever be awarded to a target, and so this property submitted to BLAST with an expectation value of is given a very high weighting to compensate for 0.01, did not return any hits. The two peaks in this fact. The other useful properties a target may E. coli O157:H7 EDL993 target scores occur for possess are, in a sense, bonuses and are scored to the same reason, except that the peak at the higher reflect this. A target does not necessarily need to score is due to genes that return no hits but have be (directly) involved in virulence in order for a been assigned some sort of function, presumably drug to neutralize an infection. However, involve- by other methods. ment in virulence may bring benefits to using a target,inthatthetargetshouldbeabsentfrommost Scoring systems and sensitivity analysis non-pathogens and also absent from humans. The The first scoring system used was designed to existence of homologues in humans does not mat- reflect what is thought to be most important in ter per se; rather, it is the similarity (or lack) of 800 700 600 y 500 c n ue 400 q e Fr 300 200 100 0 024681111122222333334444455555666667 and0 an0 an0 an0 an00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a 19d 39d 59d 79d 99nd 1nd 1nd 1nd 1nd 1nd 2nd 2nd 2nd 2nd 2nd 3nd 3nd 3nd 3nd 3nd 4nd 4nd 4nd 4nd 4nd 5nd 5nd 5nd 5nd 5nd 6nd 6nd 6nd 6nd 6nd 7 1357913579135791357913579135791 9999999999999999999999999999999 Score between Figure 2. Frequency distribution of scores for potential targets in Staphylococcus aureus, based on the first scoring systemused Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327. Assessmentofnovelantibacterialtargets 313 1200 1000 800 y c n ue 600 q e r F 400 200 0 024681111122222333334444455555666667 and0 an0 an0 an0 an00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a20 a40 a60 a80 a00 a 19d 39d 59d 79d 99nd 1nd 1nd 1nd 1nd 1nd 2nd 2nd 2nd 2nd 2nd 3nd 3nd 3nd 3nd 3nd 4nd 4nd 4nd 4nd 4nd 5nd 5nd 5nd 5nd 5nd 6nd 6nd 6nd 6nd 6nd 7 1357913579135791357913579135791 9999999999999999999999999999999 Score between Figure 3. Frequency distribution of scores for potential targets in Escherichia coli O157:H7 EDL993, based on the first scoringsystemused 1200 1000 800 y c n ue 600 q e r F 400 200 0 0 2 4 6 8 1 1 1 1 1 2 2 2 2 2 3 3 3 3 3 4 4 4 4 4 5 5 5 5 5 6 6 6 6 6 and 0 an 0 an 0 an 0 an 00 a 20 a 40 a 60 a 80 a 00 a 20 a 40 a 60 a 80 a 00 a 20 a 40 a 60 a 80 a 00 a 20 a 40 a 60 a 80 a 00 a 20 a 40 a 60 a 80 a 00 a 20 a 40 a 60 a 80 a 19 d 39 d 59 d 79 d 99 nd 1 nd 1 nd 1 nd 1 nd 1 nd 2 nd 2 nd 2 nd 2 nd 2 nd 3 nd 3 nd 3 nd 3 nd 3 nd 4 nd 4 nd 4 nd 4 nd 4 nd 5 nd 5 nd 5 nd 5 nd 5 nd 6 nd 6 nd 6 nd 6 nd 6 1 3 5 7 9 1 3 5 7 9 1 3 5 7 9 1 3 5 7 9 1 3 5 7 9 1 3 5 7 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 Score between Figure 4. Frequency distribution of scores for potential targets in Mycobacterium tuberculosis, based on the first scoring systemused the target to a human homologue which is impor- the bacterial version of a protein. In a similar way tant. Again, this is reflected in the scoring system, ‘knownfunction’and‘entryinPDB’arenotcrucial with the number of human homologues being less propertiesthatapotentialtargetmustpossess.They important than proximity. Of course the lack of simply imply that something is already known any human homologues will bring other benefits, aboutthesetargetswhichcanbeusedasajumping- such as the reduced need for QSAR studies to find off point for further investigation. ‘Copy number’ lead compounds that will selectively target only couldbepotentiallyimportantas,ifaproteinexists Copyright2004JohnWiley&Sons,Ltd. CompFunctGenom2004;5:304–327.
Description: