Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals ... PDF
Preview Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals ...
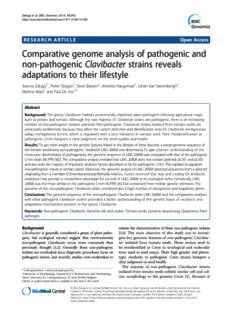
Załugaetal.BMCGenomics2014,15:392 http://www.biomedcentral.com/1471-2164/15/392 RESEARCH ARTICLE Open Access Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle Joanna Załuga1*, Pieter Stragier1, Steve Baeyen2, Annelies Haegeman2, Johan Van Vaerenbergh2, Martine Maes2 and Paul De Vos1,3 Abstract Background: The genus Clavibacter harbors economically important plant pathogens infecting agricultural crops such as potato and tomato. Although thevast majorityof Clavibacter strains are pathogenic,thereis an increasing number ofnon-pathogenic isolates reported. Non-pathogenic Clavibacter strains isolated from tomato seeds are particularly problematic becausetheyaffectthecurrentdetectionandidentificationtestsforClavibactermichiganensis subsp.michiganensis(Cmm),which is regulated with a zero tolerance in tomato seed. Their misidentification as pathogenic Cmm hampers a clear judgment on the seed quality and health. Results: Togetmoreinsightinthegeneticfeatureslinkedtothelifestyleofthesebacteria,awhole-genomesequenceof thetomatoseed-bornenon-pathogenicClavibacterLMG26808wasdetermined.Togainabetter understanding of the molecular determinantsofpathogenicity,thegenomesequenceofLMG26808wascomparedwiththatofthepathogenic Cmmstrain(NCPPB382).ThecomparativeanalysisrevealedthatLMG26808doesnotcontainplasmidspCM1andpCM2 andalsolacksthemajorityofimportantvirulencefactorsdescribedsofarforpathogenicCmm.Thisexplainsitsapparent non-pathogenicnatureintomatoplants.Moreover,thegenomeanalysisofLMG26808detectedsequencesfromaplasmid originatingfromamemberofEnterobacteriaceae/Klebsiellarelative. Genes received that way and coding for antibiotic resistancemayprovideacompetitiveadvantageforsurvivalofLMG26808initsecologicalniche.Genetically,LMG 26808wasthemostsimilartothepathogenicCmmNCPPB382butcontainedmoremobilegeneticelements.The genomeofthisnon-pathogenicClavibacterstraincontainedalsoahighnumberoftransportersandregulatorygenes. Conclusions:Thegenomesequenceofthenon-pathogenicClavibacterstrainLMG26808andthecomparativeanalyses withotherpathogenicClavibacterstrains provided a better understanding of the genetic bases of virulence and adaptation mechanisms present in the genus Clavibacter. Keywords: Non-pathogenicClavibacter,Bacterialwiltandcanker,Tomatoseeds,Genomesequencing,Quarantine,Plant pathogen Background initiatethecharacterizationofthesenon-pathogenicisolates Clavibacter isgenerallyconsideredagenus ofplantpatho- [3,4]. The main objective of this study was to investi- gens, but ecological surveys suggest that environmental, gate key genomic features of non-pathogenic Clavibac- non-pathogenic Clavibacter occur more commonly than ter isolated from tomato seeds. These strains tend to previously thought [1,2]. Generally these non-pathogenic be misidentified as Cmm in serological and molecular isolatesareoverlookedsincediagnosticproceduresfocuson tests used in seed assays. Their high genetic and pheno- pathogenicstrains.Justrecently,studieswereundertakento typic similarity to pathogenic Cmm strains hampers a clearjudgmentonseedhealth. The majority of non-pathogenic Clavibacter strains *Correspondence:[email protected] isolated from tomato seeds exhibit similar cell and col- 1LaboratoryofMicrobiology,DepartmentofBiochemistryandMicrobiology, ony morphology to the genuine Cmm [5]. Because of GhentUniversity,K.L.Ledeganckstraat35,GentB-9000,Belgium Fulllistofauthorinformationisavailableattheendofthearticle ©2014Załugaetal.;licenseeBioMedCentralLtd.ThisisanOpenAccessarticledistributedunderthetermsoftheCreative CommonsAttributionLicense(http://creativecommons.org/licenses/by/2.0),whichpermitsunrestricteduse,distribution,and reproductioninanymedium,providedtheoriginalworkisproperlycredited.TheCreativeCommonsPublicDomain Dedicationwaiver(http://creativecommons.org/publicdomain/zero/1.0/)appliestothedatamadeavailableinthisarticle, unlessotherwisestated. Załugaetal.BMCGenomics2014,15:392 Page2of14 http://www.biomedcentral.com/1471-2164/15/392 the commonbiologicalorigin(tomatoseed),highsequence strains,which could deliversomeinformative insightsinto similarities and similar physiological characteristics, the the differences in virulence determinants, genetic content non-pathogenicClavibacter strains are suggestedto be the and adaptation to a lifestyle in their natural ecological most related to Cmm. Initial in planta experiments niche(s). Genome comparison between pathogenic and demonstrated that this group of isolates is not patho- non-pathogenicstrainsbelongingtothesamespeciesisan genic to the tomato plant and they do not colonize the important and valuable approach to identify genes that vascular tissues of tomato [4]. Non-pathogenic clavi- may contribute to virulence and general fitness of the bacters neither induce a hypersensitive reaction (HR) organism. after infiltration of Nicotiana tabacum and Nicotiana In this report we present the genome analysis of non- benthamiana leaves[3],norwheninoculatedtoMirabilis pathogenic Clavibacter LMG 26808 isolated from to- jalapa (J. Van Vaerenbergh, personal communication). mato seed. The specific purposes of this study were a) to Furthermore, a majority of these strains is lacking one or generate a draft genome sequence of this strain, b) to bothCmmplasmidscarryingimportantvirulencefactors. analyze it for virulence-related gene content by compar- So far there is very little information available on non- ing it to the available genome of the pathogenic Clavibac- pathogenicClavibacterstrainsisolatedfromtomatoseeds. ter michiganensis subsp. michiganensis (Cmm) NCPPB Reports concerning the ecological niche, survival abilities 382, c) to perform a comparative analysis with the ge- or nutritional requirements are lacking. Knowledge about nomes of Cmm (NCPPB 382) [8], Clavibacter michiga- the biology of these strains is limited, not only because nensis subsp. nebraskensis (Cmn) (NCPPB 2581, released they were only recently identified as constituting a separ- withoutpublication)andClavibactermichiganensissubsp. ate Clavibacter group but also because their significance sepedonicus (Cms) (ATCC 33113) [9], pathogenic to to- in the Cmm identification procedure has not been evalu- mato, maize and potato, respectively, d) to search for ad- ated previously. Their ecological niches remain unknown; aptationstoanon-pathogeniclifestyle. routes of transmission and possible sources of these strainshavenotyetbeenrecognized. Methods High genetic and phenotypic similarities of non- StrainsandDNAextraction pathogenic Clavibacter and pathogenic Cmm strains Non-pathogenicClavibactersp. LMG26808wasreceived are the main reasons for their misidentifications as as isolate PD 5684 from Naktuinbouw, The Netherlands. Cmm in the currently recommended detection/identi- Itwasobtainedindilutionplatingonsemi-selectivemedia fication testsforCmmintomatoseeds[6].Cross-reactions accordingtothe currentmethodfordetectionofCmmin with antisera specific for Cmm and/or positive PCR reac- tomato seeds recommended by the International Seed tions with primers used for identification of Cmm illustrate Federation(ISF) [6]. LMG26808 isphenotypically similar theproximityofsurfaceantigensandgenomicsequencesof to Cmm on SCMF and CMM1T and was identified as non-pathogenic seed-borne Clavibacter to the pathogenic Cmm in commonly practiced PCR tests but showed no Cmm[3].RecentstudiesdemonstratedthatneitherPCRas- pathogenicityintomatoplants[3,4].LMG26808wasaer- saysbasedoncommonlyused16SrRNA genes or ITS re- obically grown on MTNA (mannitol, trimethoprim, nali- gion, nor those designed for the detection of known dixic acid, amphotericin) medium without antibiotics virulence factors are specific to only pathogenic Cmm at 25°C for 24 h-48 h [10]. Stock cultures were stored [3,7]. Furthermore, some non-pathogenic Clavibacter at −80˚C in MicrobankTM beads (Pro-Lab Diagnostics, strains showed fainter PCR amplicons on the gel im- Canada).Total genomic DNA was extracted according peding the correct interpretation of the results [4]. to the guanidium-thiocyanate-EDTA-sarkosyl method Taxonomically, these non-pathogenic clavibacters from described by Pitcher [11], which was adapted for Gram- tomato seeds are distinct from all Clavibacter subspe- positive bacteria by a pre-treatment with lysozyme cies (based on the analysis of housekeeping genes gyrB (5 mg/μl lysozyme in TE buffer) and incubation for and dnaA) [4]. 40 minutes at 37°C. Recent developments in the field of molecular biology and sequencingallowed generating complete genome se- quences andsubsequentlydeterminingmetabolictraitsfor Plasmidextraction many organisms. Complete genome sequences of Cmm Isolation of plasmid DNA was based on the alkaline NCPPB 382 [8], Cms ATCC 33113 [9] and Cmn NCPPB method of Anderson and McKay [12]. Agarose gel elec- 2581 (released without publication) provide genetic infor- trophoresis was performed in a Tris acetate buffer con- mation that allows for comparative studies and helps to taining 40 mM Tris, 20 mM acetic acid, and 2 mM better understand their pathogenicity characteristics and Na EDTA (pH 8.1). Gels contained 0.8% agarose and 2 host adaptation. However, no information is available electrophoresis was performed at 55 Vfor 16 hrs at 4˚C. about the genome content of non-pathogenic Clavibacter Gelswerestainedwith ethidium bromide0.5μg/ml. Załugaetal.BMCGenomics2014,15:392 Page3of14 http://www.biomedcentral.com/1471-2164/15/392 Genomesequencing Comparativegenomicanalysis Library preparation and genome sequencing was per- Artemis software was used for data management and formed by BaseClear (Leiden, The Netherlands). High- DNAPlotter was used for genome visualization [20]. The molecular weight genomic DNA was used as input for MAUVE alignment tool was used for multiple genomes library preparation using the Illumina TruSeq DNA li- sequence alignment and visualization. IslandViewer was brary preparation kit (Illumina). Briefly, the gDNA was used to analyze possible genomic islands (GI) on the fragmentedandsubjectedtoend-repair,A-tailing,ligation draft genome [21]. IslandViewer integrates two sequence of adaptors including sample-specific barcodes and size- composition GIs prediction methods, namely IslandPath- selection to obtain a library with median insert-size DIMOB [22] and SIGI-HMM [23] and one single com- around 300 bp. After PCR enrichment, the resultant parativeGIpredictionmethod,namelyIslandPick[21]for library was checked on a Bioanalyzer (Agilent) and genomicislandprediction. quantified. The libraries were multiplexed, clustered, ISsaga application from ISfinder server [24] was used and sequenced on an Illumina HiSeq 2000 with paired- to identify insertion sequences (IS) and transposons in end 50 cycles protocol. The sequencing run was analyzed the draft genome of LMG 26808. Sequences exhibiting with the Illumina CASAVA pipeline (v1.8.2). The raw se- homology to IS and transposon sequences were verified quencing data produced was processed removing the with the Mobilomics software [25]. The core genome sequence reads which were of too low quality (only was estimated using the Phylogenetic profiler tool that is “passing filter” reads were selected) and discarding reads part of the IMG system (https://img.jgi.doe.gov/cgi-bin/ containingadaptor sequences orPhiXcontrol withanin- er/main.cgi)at a similaritycutoffof50%. housefilteringprotocol. The presence of possible virulence-related genes and Apaired-end(PE) DNAlibrarywith amean insert size genes expressed during tomato infection in the draft of 300 bp was sequenced with average reads of 101 bp genome of the non-pathogenic Clavibacter was analyzed onanIlluminaGenomeHiSeq2000(IlluminaInc.).Next, by comparing it with tomato pathogen Cmm NCPPB a mate-paired (MP) DNA library with a mean insert size 382. The comparative screening of the gene content was of 3800 bp was sequenced with average reads of 51 bp performed in RAST, IMG-ER and EDGAR [26]. Absence on an Illumina Hiseq2000 (Illumina Inc.). Automatic or presence of coding sequences in each genome, as re- trimming (based on a threshold of Q=20) and assembly ported by RAST, IMG-ER and EDGAR were independ- was performed using CLC Genomics Workbench v5.0. ently confirmed by performing protein and nucleotide An initial de novo assembly was performed in CLC Gen- BLASTqueriesinthetargetgenomes.Proteinswithamino omics Workbench v5.0 using the quality trimmed and acid sequence similarities higher than 50% and with a paired readsfromthePEandMPreads.Allcontigsshorter coverage higher than 70% were considered homologs. than 200 bp were discarded. Remaining N-nucleotides in Based on the RAST, IMG-ER and EDGAR annotation re- the scaffolds, introduced during scaffolding, were removed sults,the presence of known and putative virulence fac- from the final sequence by breaking up the scaffolds tors, pathogenicity related genes and genes uniquely back into contigs where they were encountered. The present in the non-pathogenic Clavibacter LMG 26808 quality of the final draft genome sequence was com- were investigated. Identification of orthologous groups pared to the initial PE-based de novo assembly through between fouravailableClavibactergenomeswasachieved comparative read-mapping in CLC Genomics Work- by OrthoMCL analyses [27]. OrthoMCL clustering ana- bench v5.0 using the trimmed read sets. Contigs were lyseswere performed using default parameters with the ordered automatically with MAUVE [13] and manually P-value Cut-off=1×10−5. with Artemis [14] by comparing with Cmm NCPPB 382. Deposition The current draft genome sequence was deposited at Genomeannotation Genbank under accession number AZQZ00000000 after Functional annotation and metabolic reconstruction were automatic annotation by the PGAAP online annotation performed with (1) the Rapid Annotation Subsystem pipeline. Technology (RAST) server [15], using Glimmer [16] for genecallingandallowingframeshiftcorrection,backfilling Results and discussion of gaps and automatic fixing of errors, with (2) the Inte- Generalfeaturesofnon-pathogenicClavibacterLMG grated Microbial Genomes Expert Review (IMG-ER) an- 26808 notation pipeline (https://img.jgi.doe.gov/cgi-bin/er/main. Genome assembly using paired-end and mate-paired cgi) [17]. Assigned functions were checked with BLAST reads resulted in a 3.47 Mb sequence represented in 70 [18].Alignmentandphylogeneticanalysiswereperformed contigs from which the longest covered more than one withMEGA5.0[19]. million bp (Table 1). The initial PE de novo assembly Załugaetal.BMCGenomics2014,15:392 Page4of14 http://www.biomedcentral.com/1471-2164/15/392 Table1Genomecharacteristicsofthenon-pathogenic couldbedetected.TheGCcontentofthedraftgenomeav- ClavibacterLMG26808 erages 72%. There are 46 tRNA genes and two complete Genomecharacteristics Non-pathogenic rRNA operons. A total of 3218 protein-coding genes are ClavibacterLMG26808 predicted in non-pathogenic Clavibacter strain (in IMG- No.contigs(>200bp) 70 ER),whichissimilartotheCmmgenomeNCPPB382that Totalcontigsize(bp) 3,476,455 contains 3107 protein-coding genes. The genome of the non-pathogenicClavibacterstraincontains685(21.3%)pro- N50(bp)afterscaffolding 383,456 teins without predicted function being either annotated as Largestcontigsize(bp) 1,028,177 conserved hypothetical proteins or proteins with unknown GCcontent(%) 72.01 function. No.RNAcalls 7rRNA The number of genes detected in the draft genome of 46tRNA LMG 26808was higher thaninthe otherthree complete No.CDScalls 3218 Clavibacter genomes (Table 2). The average nucleotide identity (ANI) between the draft genome of the non- NCBIaccessionno. AZQZ00000000 pathogenic Clavibacter and the three published Clavi- Numberofinsertionelements 10 bacter genomes Cmm NCPPB 382 (NC_009480.1), Cms ATCC 33113 (NC_010407.1) and Cmn NCPPB 2581 wasusedforscaffoldingwiththeMPdataset.Inthefinal (NC_020891.1) was determined using the in silico DNA- consensus sequence each base matched at least Phred DNA hybridization (DDH) method included in the soft- quality scoreof35.LMG26808containsonechromosome ware JSpecies [28]. The results indicated that LMG andevidenceofapresenceofaplasmidthatshowedahigh 26808 is genetically most related to Cmm NCPPB 382 similarity to a Klebsiella pneumoniae Kp11978 plasmid (94.96% ANI), followed by Cmn NCPPB 2581 (92.75% pOXA-48 (JN626286.1). The genesofKp11978were found ANI) and Cms ATCC 33113 (92.48% ANI). Although on 15 contigs in a draft genome of LMG 26808 (estimated based on the ANI values the LMG 26808 genome is the sizeofthesecontigs~48kbp,%GC~50%)(Additionalfile1: most similar tothatofpathogenicCmmNCPPB382,the Table S1). No sequences of known Clavibacter plasmids synteny plots of LMG 26808 and Cmn NCPPB 2581 and Table2Comparisonofgenomecharacteristics(basedonIMG-ERserver) GenomeName Clavibactermichiganensis Clavibactermichiganensis Clavibactermichiganensis Non-pathogenic subps.michiganensis subsp.sepedonicus subsp.nebraskensis ClavibacterLMG26808 NCPPB382 ATCC33113 NCPPB2581 Accessionnumbera NC_009480.1 NC_010407.1 NC_020891.1 AZQZ00000000 Host tomato potato maize * Disease bacterialwiltandcanker potatoringrot wiltandblight non-pathogenic Size 3395237 3403786 3063596 3476455 Genes 3169 3168 2936 3282 CDS 3107 3117 2890 3218 CDS(%) 98.04 98.39 98.43 98.05 RNA 62 51 46 64 rRNA 6 6 6 7 tRNA 45 45 30 46 Enzymes 759 712 740 750 CRISPR 1 0 0 1 GC% 72 72 73 72 Codingbases 3041059 2955244 2823671 3074588 Signalpb 281 234 219 140 Signalp(%) 8.87 7.39 7.46 4.27 HomologstoLMG26808(%)c 2716(87.4) 2457(78.8) 2531(87.5) - aOnlytheGenbankrecordsofthechromosomesaregiven. bNumberofgenescodingsignalpeptides. cCalculatedusingaGenomeGeneBestHomologstoolincludedinIMG-ERserver. *Isolatedfromtomatoseeds,hostunknown. Załugaetal.BMCGenomics2014,15:392 Page5of14 http://www.biomedcentral.com/1471-2164/15/392 the percentage of homologous genes shared by LMG amongClavibacter strains most likely contribute to the 26808 and Cmn NCPPB 2581 are also considerably high differences inthebacteriallifestyle.Phage-relatedrecom- (Table 2, Figure 1). The genomes of LMG 26808, NCPPB binases (e.g. Cl_00892, Cl_03056), integrase/resolvase 382 and NCPPB 2581 are collinear with less than 5 re- (e.g. Cl_02713) and other insertion elements (transpo- combinationalbreakpoints. sases, e.g. Cl_03190) associated with a phage were found Comparison on a genomic scale revealed a high con- in higher numbers in the genome of LMG 26808 than in servation in the gene sequence among genomes of LMG the Cmm NCPPB 382 genome (Table 3). The genome of 26808, NCPPB 382 and NCPPB 2581 (Figure 1). There LMG 26808 contained sixteen genes coding for transpo- are 299 genes (~10%) present in the LMG 26808 draft sasesandrecombinases(Table3).Thisnumberwasmuch genome that were not detected in the Cmm NCPPB 382 lower in comparison to more than 100 genes found in genome. Forty eight of them were detected in Cmn and/ Cms (ATCC 33113)[9]. Detected IS belonged to IS3,IS4, or Cms genomes (Additional file 1: Table S2). 37 unique IS5, IS6 and IS1380 families. Transposases were repre- genes of LMG 26808 were associated with the plasmid sented by Tn3 (20%). No pseudogenes among transpo- and/or low GC regions. 214 of the unique genes were sases and recombinases were detected suggesting that foundinthecorechromosomeofLMG26808(Additional these elements may encode functional genes.None of the file 1: Table S2). Almost half of the genes specific for IS elements found in LMG 26808 has homologs in the LMG26808belonged to hypothetical or unknown pro- other Clavibacter strains. The most common IS element teins (120). Remaining sequences were coding for add- in Cms ATCC 33113 is IS1121 [9]. Cmm NCPPB 382 itional ABC transporters, antibiotic resistance genes, containsonlyafewIS,whicharemostprobablynonfunc- acetyltransferases and several enzymes that in majority tional [8]. Cmn NCPPB 2581 contained only two types of couldnotbeassignedtoanyKEGG(KyotoEncyclopediaof IS,namelyILSre2andISNCY(predictedbyISsaga). GenesandGenomes)pathway(Additionalfile1:TableS2). The comparison of functional categories as defined When compared to other Clavibacter genomes, LMG by COG (Clusters of Orthologous Groups) showed no- 26808 appeared to not have experienced gene losses and ticeable differences in the gene content in categories of despiteitisconsideredonlyadraft,themajorityofimport- ‘carbohydrate transport and metabolism’ and of ‘trans- antgenesinvolvedinbasicmetabolismandgeneregulation lation, ribosomal structure and biogenesis’. All included could be detected. Comparative analysis (based on KEGG Clavibacter strains contained a higher percentage of pathways) showed that LMG 26808 lacks sulfate and ni- genes in these two categories than a free-living organ- trate reduction pathways suggesting that its capability of ism Escherichia coli 081 ED1a or a tomato pathogen as survival in soli might be similar to this observed in Cmm e.g. Pseudomonas syringae pv. tomato T1 (Additional NCPPB 382. The core genome consists of 2316 homologs file 1: Figure S1). These observations are supporting the found in all four Clavibacter genomes. LMG 26808 con- hypothesis that compared Clavibacter strains can most tains 12 genomic regions exhibiting a lower GC content probably utilize different sugars as an energy source and (Additional file 1: Table S3). Several genes coding for pro- that they possess a wide range of transport systems that teins within these regions were found to contribute to the enable the efficient trafficking of the substrates and prod- fitness of the bacterium (Cl_02679 coding for ABC-type ucts. The presence of a high number of genes involved in Fe3+-siderophore transport system; Cl_03044 coding for translation, ribosomal structure and biogenesis implies permease component, chloramphenicol acetyltransferase thatthesebacteriarespondmoreeffectivelyandrapidlyto (EC 2.3.1.28); Cl_03094 coding for multidrug-efflux trans- nutritional resources, which can be an important advan- porter). The genome heterogeneity and genetic diversity tageinachangingenvironment. Figure1Syntenicdotplotsshowingthesimilarityofthegenomesincludedintheanalysis.A)non-pathogenicClavibacterLMG26808(x-axis) andCmmNCPPB382(y-axis);B)non-pathogenicClavibacterLMG26808(x-axis)andCmnNCPPB2581(y-axis);C)non-pathogenicClavibacterLMG 26808(x-axis)andCmsATCC33113(y-axis);D)CmmNCPPB382(x-axis)andCmnNCPPB2581(y-axis)(DiagramsgeneratedinIMG-ER). Załugaetal.BMCGenomics2014,15:392 Page6of14 http://www.biomedcentral.com/1471-2164/15/392 Table3MobilegeneticelementsfoundinthegenomeofLMG26808(BasedontheannotationresultsfromIMG-ER, RASTandEDGAR) CDSidentifiers COG COGannotation Pfam Position Length(bp) Cl_00892 COG4974 Site-specificrecombinaseXerD,phage_integrase pfam00589 Contig3(50199to51350) 1152 Cl_00935 COG4974 Site-specificrecombinaseXerD,phage_integrase pfam00589 Contig5(8554to9540) 987 Cl_01562 COG4974 Site-specificrecombinaseXerD,phage_integrase pfam00589 Contig5(682570to683556) 987 Cl_01811 COG1842 PhageshockproteinA(IM30),suppresses pfam04012 Contig5(938633to939376) 744 sigma54-dependenttranscription Cl_01968 COG3600 Uncharacterizedphage-associatedprotein pfam13274 contig7(7492to7956) 753 Cl_03043 COG4679 Phage-relatedprotein pfam05973 contig15(974to1303) 330 Cl_03056 COG4974 Site-specificrecombinaseXerD,phage_integrase pfam00589 contig15(22760to23485) 726 Cl_03252 COG4974 Site-specificrecombinaseXerD,phage_integrase pfam00589 contig46(189to866) 678 Cl_02713 COG2452 Predictedsite-specificintegrase-resolvase pfam12728 contig11(350076to350522) 447 Cl_03047 COG2801 Transposaseandinactivatedderivatives,Tnp1,IS3_IS150 pfam01527 contig15(9545to11077) 1533 Cl_03189 - TransposaseDDEdomain,Tnp1,IS1380 pfam01609 contig28(1417to2532) 1116 Cl_03190 COG4644 Transposaseandinactivatedderivatives,TnpAfamily,Tn3 pfam01526 contig28(4206to5753) 1530 Cl_03209 COG4644 Transposaseandinactivatedderivatives,TnpAfamily,Tn3 pfam01526 contig33(10984to12801) 1818 Cl_03210 COG3316 Transposaseandinactivatedderivatives,IS6 pfam13610 contig33(12848to13552) 705 Cl_03211 COG2801 Transposaseandinactivatedderivatives,IS3_IS150 pfam13276 contig33(14010to13498) 513 Cl_03212 COG2963 Transposaseandinactivatedderivatives,Tnp1,IS3 pfam01527 contig33(14489to14343) 147 Cl_03214 COG3316 Transposaseandinactivatedderivatives,IS6 pfam13610 contig34(391to116) 276 Cl_03235 - Transposase,Tnp1,IS5_IS903 pfam13737 contig39(1373to2212) 840 Cl_03261 - TransposaseDDEdomain,Tnp1,IS4 pfam01609 contig51(1to1188) 1188 Cl_03204 - Mobileelementprotein - contig33(530to366) 165 peg.1244 - Mobileelementprotein - contig28(149to742) 594 Cl_03063 - Mobileelementprotein - contig15(32765to34252) 1488 peg.807 - Resolvase-like - contig15(6452to6847) 396 peg.1245 - Tn1transposase - contig28(741to1088) 348 Cl_03045 - Gifsy-2prophageprotein - contig15(7251to7544) 294 Cl_03251 - putativebacteriophageprotein - contig45(3200to4171) 972 Cl_01918 - elementsofexternalorigin;phage-related - contig6(14793to15281) 489 functionsandprophages Genomicislands regionsaccountsfor265kb(~7%ofthegenomesize).The The analysis of the LMG 26808 genome showed that at equivalent of PAI of Cmm NCBI 382 (130 kb) containing least12regionswithlowerGCcontentsdistributedamong two regions chp and tomA with important genes respon- different contigs could be distinguished (Additional file 1: sible for effective plant colonization, was not found in TableS3).Partsofgenomicislands3and4foundinLMG LMG 26808, nor in other Clavibacter genomes. However, 26808 partly overlap with the chp region of pathogenicity a number of orthologswerefound in all three Clavibacter island (PAI) described previously in Cmm NCPPB 382 genomes (Additional file 1: Table S4). The higher number (Additional file 1: Table S3). Genomic islands with lower of orthologs of genes encoded on chp and tomA regions GC% are thought to be integrative elements that exhibit (as detected by OrthoMCL) was found in Cms ATCC different codon usage relative to the rest of the genome, 33113(32),followedbyLMG26808(17)andCmnNCPPB encodefortransposases, integrases and aretypicallyfound 2581(10). Onlysix orthologs of PAI (chpregion) found in attRNAloci.Theiracquisitionismostlyaresultofactions LMG 26808 were located on the low GC region 3 and 4 of phages, transposons or horizontal gene transfer [29]. (Additionalfile1:TableS3). SomeofthegenespresentintheseregionsinLMG26808 Genomic regions with lower GC content can contain were detected previously in the genome of Cmm NCPPB diverse genes exhibiting functions in many metabolic 382 but the majority represents regions that were not processes. The longest region found in LMG 26808 found in Clavibacter subspecies. The total size of these (more than 100 kb) included genes coding for antibiotic Załugaetal.BMCGenomics2014,15:392 Page7of14 http://www.biomedcentral.com/1471-2164/15/392 resistance (beta-lactamase class A, Cl_03208, Cl_03230), neededtoelucidatetheexactfunctionsofthesegenes.The transposases (Cl_03209, Cl_03212) and many hypothet- smaller plasmid pCM1 was not detected during the plas- ical proteins(Cl_03223,Cl_03183)someofwhichshowed mid extraction, nor were its sequences found in the gen- the highest similarity on the protein level to genes found ome sequence of LMG 26808. Despite that two DNA on Klebsiella pneumoniae plasmids. Genomic region 3 fragments of LMG 26808 showed to be highly similar to containssomegenes that showed similarities tothe genes two plasmid-encoded genes, namely pCM1_0018 and foundinpCM2plasmidofCmmNCPPB382.Themajor- pCM1_0020, the reciprocal BLAST search revealed that ity of them are hypothetical proteins and two of them these sequences from the non-pathogenic Clavibacter codeforacetyltransferasesAdditionalfile1:TableS3.Pre- genome are more similar to the chromosomally encoded vious studies indicated that some pathogenic Clavibacter CMM_1065 and CMM_2443, respectively. Interestingly, strains lacking pCM1 and pCM2 showed a positive signal the latter gene encodes CelB, which is a putative secreted inhybridizationexperimentswithspecificplasmidregions cellulase that contains a cellulose-binding domain (endo- of Cmm NCPPB 382 implying that some of the genes 1,4-beta-glucanase). Chromosomally encoded celB misses found originally in Cmm plasmids may be actually one of three protein coding domains present in the celA chromosomally-encodedinotherCmmstrains[30]. gene. The missing endoglucanase C-terminal domain is Genomic regions 7, 9 and 10 with lower GC content similar to the α-expansin protein family that occurs in contained some genes encoding transposases and recom- plants and is essential for development of wilting and for binases, which might imply their possible exchange/ degradationofcrystallinecellulose[8,32].Thelackordis- mobilization ability. In region 7 one phage-related gene ruption of any of these domains of celA inevitably led to (Cl_03043), showing homology to prophage protein gp49, the disability to induce disease symptoms in a tomato was detected. Its presence may represent the remains of plant[32]. prophagegenes. Surprisingly, the genome analysis showed the presence of sequences found in Klebsiella pneumoniae plasmid Plasmidcontent pOXA-48 (61881 bp). The presence of sequences from a LMG 26808 did not contain any of two known virulence plasmid of Gram-negative bacteria in a Gram-positive plasmids found in pathogenic Cmm NCPPB 382. How- Clavibacter strain is rather unusual and has not been re- ever, the plasmid extraction demonstrated the presence ported previously. Although the genome sequence of of one plasmid, which size was slightly smaller than that LMG 26808 is only a draft and therefore incomplete, we of plasmid pCM2 (70 kb). Initially, we assumed that it could not detect any sequences that could be attributed might be a pCM2 that lost some genes because in the toaKlebsiellapneumoniaeKp11978chromosome. previous study we could not detect the presence of the Theexchange ofgeneticmaterial between various pro- pat-1 gene, which is encoded on the Cmm plasmid [4]. karyotes is well known and has been extensively studied Even though we did not detect the complete pCM2, over the last few decades [33-37]. Although it was dem- some of the genes originally encoded on this plasmid onstratedforbacteriathatthegeneexchangeisobserved were found in LMG 26808 (Additional file 1: Table S5). more frequently between closely related genera with a Except for two genes involved in the putative conjugal similar GC content and exhibiting high sequence simi- transfer (pCM2_0013 and pCM_0019, coding for TraA larities there are examples of recent gene transfers be- and TraG, respectively), all the remaining genes showed tween distantly related bacteria (e.g. Actinobacteria and to code for hypothetical or putative secreted proteins. gammaproteobacteria) [38]. All of them were detected on contig 6 but the order in Conjugational transfer is considered the most efficient which they were found in LMG 26808 did not match way of LGT [39,40] that contribute the most to the the order demonstrated in pCM2. Moreover, there are spread of antibiotic resistance among different bacteria more genes present on contig 6, some of which showed [41]. This type of LGT is widely encountered among to be homologous to proteins from the Cmm chromo- various bacterial species and even between bacteria and some (Cl_01961-Cmm_02708, Cl_01957-Cmm_01374). Archaea [42] on the one hand and between bacteria and These observations may suggest that some of these plas- higher organisms such as Saccharomyces cerevisiae [43], mid genes were incorporated in the genome of LMG or plants [44] on the other hand. Conjugational plasmid 26808. exchange was also observed within the genus Clavibac- The observation that some genes from pCM2 that ter in which the endophytic CMM100 strain (cured of were expressed during tomato infection by Cmm [31] pCM1 and pCM2 plasmids) was able to acquire these had orthologs found in LMG 26808, might suggest that plasmids from other pathogenic Cmm strains and re- although their function is unknown, they may be essential store pathogenicity [45]. Furthermore, transformation fornon-pathogenicClavibacteraswellaspathogenicCmm experiments carried out with Clavibacter xyli subsp. strains(Additionalfile1:TableS5).Furtherinvestigationis cynodontis (currently reclassified to the genus Leifsonia) Załugaetal.BMCGenomics2014,15:392 Page8of14 http://www.biomedcentral.com/1471-2164/15/392 demonstrated the possibility to acquire an IncP plasmid factors in recent reports [30,31,52,53]. The genome of fromEnterobacteriaceaebythisGram-positiveActinobac- LMG26808didnotcontainthemostprominentvirulence teria, which provided another evidence of conjugational factors pat-1 and celA. Their absence may be directly transferbetweendiversetaxa[46]. linkedwiththeabsenceofthepCM1andpCM2plasmids Klebsiella pneumoniae strains were found in many im- in the non-pathogenic Clavibacter. However, southern portant crops such aspotato, maize, soybean, cotton and hybridization experiments with plasmid fragments con- tomato[47,48].Manyofthesestrainscarryplasmidsthat taining virulence factors showed that in some plasmid- contain antibiotic resistance genes and possess the conju- freepathogenicCmmstrainsthesevirulencedeterminants gationtransfersystemswhichenablethegenemobilization hadhomologuesonthechromosome[30]. and exchange among and outside Enterobacteriaceae and A proteomic study of tomato-Cmm interaction identi- other bacterial genera [49]. Some genes encoded on the fiedseveralbacterialproteinswithaputativeroleinsignal Klebsiellapneumoniaeplasmidsexhibithighsimilaritiesto perception, transduction and response to impulses. They regions found previously in Escherichia coli and Yersinia belongtotwo-componentsystemproteins,transcriptional genomes,implyingthatthereisanactivegeneticexchange regulators and other DNA binding proteins. They are be- amongstrainsofthesegenera[50]. lieved to play a role in sensing the tomato plant environ- Although an acquisition by LMG 26808 of a relatively ment and initiating pathways, possibly leading to disease large plasmid originating most probably from a member development [31]. All putative genes encoding proteins of Enterobacteriaceae/Klebsiella relative by LMG 26808 thatare probablyinvolved in signalexchange betweento- (Additional file 1: Figure S2) was unexpected and unpre- matoandbacteriumcouldbeidentifiedinthegenomese- cedented, a similar occurrence was previously reported quenceofLMG26808(Additionalfile1:TableS6). by Baltrus and coworkers. They detected a recent acqui- As a non-pathogenic bacterium, LMG 26808 was hy- sition of a megaplasmid by two cucumber isolates of pothesizedtocontainlessgenomicinformationforhydro- Pseudomonaslachrymans [51]. It was suggested that this lytic enzymes that are known to be expressed during acquisition resulted from an important ecological shift tomato infection with Cmm [31]. As expected, the most across closely related Pseudomonas members and that important group containing genes coding for secreted the plasmid-encoded genes may be advantageous for the proteases from Pat-1 family was largely absent in LMG recipientbacteria. 26808 (Additional file 1: Table S6). Additional pat-1 AsKlebsiellapneumoniaeandClavibacterstrainsthrivein homologues encoded on the pCM2 plasmid (plasmid thesameenvironmental niche (associated withtomato)and homologs of pat-1, phpA and phpB) were also absent. because of examples of possible genetic material exchange From seven genes encoding putative serine proteases between distantly related bacteria we can hypothesize that chpA-chpG (chromosomal homologs of pat-1) [54] only theacquisitionofplasmid sequencesencodingantibioticre- sequences similar to chpF and chpG were detected (chpF sistancegenesmightprovideacompetitiveadvantageforthe and chpG are orthologs with nucleic acid sequence simi- non-pathogenicClavibacterstrainLMG26808. larity of 69.1% and amino acids sequence similarity of 68%). Both these sequences, however, matched to the Non-pathogeniclifestyle same region and a reciprocal best BLAST hit confirmed Non-pathogenic Clavibacter strains from tomato seeds the presence of only chpF. Interestingly, the lack of chpG tested in the previous study [4] as well as other strains may be a possible explanation for the disability of LMG tested by Jacques and coworkers [3] did not introduce 26808 to produce a HR in nonhost plants since the chpG any disease symptoms when tested on tomato plants. mutant in Cmm was unable to cause an HR in Mirabilis Possible explanations for the non-pathogenic nature of jalapa [8]. Moreover, the low colonization efficiency of these strains are i) the lack of two plasmids present in LMG 26808 could be attributed to the lack of the chpC pathogenic Cmm and carrying virulence factors, ii) the gene. A chpC mutation in the pathogenic Cmm NCPPB absenceofthepathogenicityisland andiii)a significantly 382resultedinadrasticreductionincolonizationabilities lower number of genes coding for extracellular hydrolytic in tomato plants [8,55]. Pseudogenes chpA, chpB and enzymesincludingseveralimportantserineproteases,glyco- chpD were not found in LMG 26808. The family of chp sylhydrolasesandtheplantcellwall-hydrolyzingenzymes. genes is important for plant-pathogen interaction in In pathogenic Cmm, main virulence factors cel-A and Cmm, but probably also in Cms where four orthologs pat-1, encoded on pCM1 and pCM2, respectively, are re- were found. Cmn genome had no orthologs of these quiredtoinducediseasesymptoms(wiltingandcanker)in genes. tomatoplants[52,32].Moreover,genescodingforthepro- The majority of members of secreted serine proteases duction of extracellular enzymes, such as endoglucanase, of the Ppa family (PpaA-PpaJ) that are encoded in sev- polygalacturonase,xylanase,serineproteasesandotherse- eral different loci on the chromosome and on pCM1 cretedproteinshavebeenimplicatedaspossiblevirulence could not be found in LMG 26808. Orthologs of ppaI Załugaetal.BMCGenomics2014,15:392 Page9of14 http://www.biomedcentral.com/1471-2164/15/392 and ppaF were found in LMG 26808. Cms ATCC 33113 LMG 26808 suggesting that their function might not be contained orthologs of ppaB1, ppaB2, ppaF, ppaI, ppaA restricted to disease development alone. The glycosyl hy- in the chromosome and ppaC on pCS1 plasmid. On the drolasesarenotconsideredas bonafide virulencefactors, contrary Cmn NCPPB 2581 had only one ortholog of but as reflecting the adaptation to the differing compos- ppaF. Since ppaA and ppaC genes are important for ition of nutrients in planta allowing the survival inside of plant colonization [8] and they were absent in LMG theplant. 26808, it can be another evidence why non-pathogenic Very important functions involved in transport and Clavibacter strains are poorly colonizing tomato plants. metabolism are linked to the presence of ABC and other Indeed, secreted serine proteases studied in pathogenic transporters that ensure the uptake of amino acids, Cmm are thought to presumably facilitate the inter- metals, sugars, oligopeptides, etc. Some of these trans- action between Cmm and its host plant and are believed porters that were expressed during tomato infection by toplayafunctioninpathogenicitybyapossibleutilization Cmm may utilize plant molecules to support its metab- of plant proteins [31]. Their lack might imply that inter- olism. ThegenesfoundinthegenomeofLMG26808that action between LMG 26808 and tomato is actually very codefortransportersthatareknowntobeexpresseddur- limited. TomA gene of Cmm NCPPB 382 (CMM_0090), ing plant infectionby Cmm are listed in Additional file 1: encodingendo-1,4-betagalactosidaseinvolvedindetoxifi- Table S7. Interestingly, only five orthologs of fifty seven cation of the alfa-tomatine, had orthologs in three other transporters could not be found in LMG 26808. Further- Clavibacter genomes. However, the similarity based on more, the genome of non-pathogenic Clavibacter con- the amino acid sequence was rather low (coverage (%)/ tainedadditionaltransportersthatwerenotpresentinthe identity (%): 47/24 in Cms, 47/23 in Cmn and 47/22 in pathogenic Cmm genome (Additional file 1: Table S2). LMG26808). Some of them are supposed to play a role in the active Genes coding for subtilases sbtA, sbtB and sbtC are drug transport and cell protection from toxic metabolites known to be secreted during the plant infection [31]. (C_03094 and Cl_03219). Another very important ex- Orthologs of these three subtilases genes were found in ampleofadditional ABC transporters in the genome of all four Clavibacter genomes. Sbt proteins of Cmm are LMG 26808 (not found in other three Clavibacter ge- highly similar to different tomato subtilases, some of nomes) aretransportersinvolvedinironcomplextransport which have been associated with wound formation and (ABC-typeFe3+siderophoretransportsystemCl_02679and pathogen responses [56] and may play a role in the dis- ABC-type cobalamin/Fe3+−siderophores Cl_ 02677) ease development. Because they are present in the non- (Additional file 1: Table S8). An alternative iron uptake pathogenic Clavibacter strain their function probably system found in LMG 26808 might be advantageous in cannot be solely associated with the disease develop- an iron deficient environment. This data suggests that ment. Cellulases and pectinases are the most important LMG 26808 is probably able to utilize a broad variety enzymes degrading plant cell walls. In many bacteria of compounds to maximize its survival changes. Many genes encoding these enzymes were found to be viru- environmental bacteria were shown to contain a high lence determinants [57]. In the genome of LMG 26808 number of transporter genes in support of an environ- genes for pectate lyases, pelA1 and pelA2 and cellulase mental lifestyle [60]. celA were not found. However, another cellulase celB, Observations described above correlate well with the the polygalacturonase pgaA (whose substrate is pectin), initial assumptions that suggested that non-pathogenic xysA (whose substrate is β-1,4-xylan) and an arylesterase Clavibacter strains must have lost or never contained (which hydrolyzes ester bonds between hemicelluloses prominent virulence determinants responsible for dis- and lignin) [58] were present in LMG 26808 (Additional ease induction in tomato plants. These hypotheses were file 1: Table S6). These findings support the thesis that partially underpinned by similar findings in another draft the non-pathogenic Clavibacter strain is probably less genomeofnon-pathogenicClavibacterLMG26811,which efficient in digesting pectins and cellulose into simpler lacks the majority of virulence factors including two main by-products than the pathogenic Cmm that is equipped determinants.Italsocontainedlessplantcelldegradingen- with manyvariousplant celldegradingenzymes. zymes than pathogenic Cmm NCPPB 382 (data not Enzymes from a large group of glycosyl hydrolases shown). Furthermore, the comparative genome analysis (GH) which hydrolyze the glycosidic bond between two of LMG 26808 and Cmm NCPPB 382 revealed that carbohydrates or between a carbohydrate and a noncar- some putative virulence factors, determined based on bohydrate molecule[59]werealsoexpressedduringplant expression levels obtained from the proteomic study of infection of Cmm. Therefore, many of them are assumed tomato-Cmm interaction [31], were also present in to be potential virulence factors that can hydrolyze sub- LMG 26808, which may indicate their redundant func- strates of plant origin [31]. Ourresults demonstrated that tions and suggest that they are not critical for Cmm genes for the majority of these enzymes are present in virulence. Załugaetal.BMCGenomics2014,15:392 Page10of14 http://www.biomedcentral.com/1471-2164/15/392 Survivalintheenvironmentalniche and YoeB T-A genes were found in many bacterial ge- Non-pathogenic Clavibacter strain LMG 26808 was iso- nomes and sometimes more than one copy per genome lated from tomato seeds yet knowledge on its environ- [63]. It was demonstrated that T-A systems are present mental niche is largely lacking. Ecological niche(s) and onlyinenvironmentalandfree-livingorganismsandwere transmission routes have not yet been found. Prelimin- not detected in intracellular bacteria [63]. The BLASTp ary results with colonization experiments showed poor analysis of YefM and YoeB genes from LMG 26808 re- colonization of vascular tissues of tomato and seemingly vealedhighsimilaritiestoproteinsfromRhodococcuspyri- lower survival potential of LMG 26808 in comparison to dinivoransAK37andMicrobacteriumtestaceumStLB037, Cmm [4]. The HR was not induced in Mirabilis jalapa respectively. The T-A system found in LMG 26808 was (J. Van Vaerenbergh, data not published), indicating that not present in the pathogenic Cmm,but YefM(peg.1235) non-pathogenic Clavibacter strains do not contain genes was present in another non-pathogenic Clavibacter strain that would be recognized by the plant to trigger the ac- LMG 26811 (data not shown). Interestingly, Cmn con- tive plant defense response. tained another putative toxin-antitoxin system. T-A sys- Antibiotic resistance. In the genome of LMG 26808 temsarenotessentialfornormalcellgrowth,nevertheless several additional genes responsible for antibiotic resist- theyarepresentinmanybacteriaandArchaea[63].Based ance were detected (Additional file 1: Table S2). They on the frequency of T-A systems, it was suggested that coded for beta-lactamases (Cl_03263, peg.1233, peg.1766, they play subtle roles that are advantageous for cell sur- peg.1776), chloramphenicol acetyltransferase (Cl_03044) vivalintheirnaturalhabitats.Toxinsmayfacilitatecellular and tetracycline efflux protein TetA (peg.1764). They adaptation of an organism to changing environments by showed the highest similarity togenes found in Klebsiella slowingdownits cell growth,inhibitingits cell growth,or pneumonia, Escherichia coli and Salmonella enterica causing some ofits cells to die [64]. Itis possible that the suggesting that they could originate from these bacteria. presence of a T-A system in the LMG 26808 genome In addition to the above genes, the genome of LMG increases the fitness of this bacterium in the occupied 26808 contains two drug efflux transporters (Cl_03219, environmental niche. Differences in the detected toxin- Cl_03094) not found in pathogenic Cmm NCPPB 382. antitoxin systems in particular Clavibacter subspecies Interestingly,LMG26808containedglyoxalase/bleomycin might be attributed to different ecological niches and resistance protein (Cl_03100), which probably constitutes inhabited hosts. the resistance to bleomycin-antibiotic produced by some ErrorproneUmuDCoperon.SOSmutagenesisresponse Streptomycesstrains[61]. inbacteriaincludeserror-proneanderror-freeDNAdam- Thepresenceofadditionalacetyltransferasesmightsug- age repair responses that are activated after exposure to gest that LMG 26808 exhibits broad resistance to certain different antibiotics, chemical compounds or radiation antibioticsassomeoftheacetyltransferases(GNATsuper- [65]. In Escherichia coli UmuDC proteins are involved in family)catalyzetheselectiveacetylationofoneofthefour error-pronebypassofUVlesionsandUmuCproteinspos- amino groups found on a diverse set of aminoglycosides sess DNA polymerase activity. In the SOS process many withantibioticproperties[62].Acetylationreducestheaf- genesgetinducedandtheirproductsareinvolvedinDNA finity of these compounds for the acceptor tRNA site on repair, replication and cell cycle control in order to repair the 30S ribosome. As a consequence, bacteria expressing the DNA damage [66]. The genes coding for this operon these genes are resistant to some aminoglycosides with were found in LMG 26808 and also in another non- antibiotic properties. The ability to cope with antibiotics pathogenic Clavibacter, LMG 26811 (data not shown), producedbyorganismswithwhichnon-pathogenicClavi- implying that their cells might have higher abilities to bacterstrainsshare the environmental niche is a signifi- recover after exposures to UV and/or other types of cant adaptive advantage. The growth of Clavibacter chemicals retrieved during the seed certification proce- strains in culture is often inhibited by other faster grow- dures. Some of the sequences coding for genes of ing organisms. Therefore, the presence of genes coding UmuC operon (e.g. peg.1211) and antibiotic resistance for antibiotic resistance might be the reason why non- genes (e.g. Cl_03263) described above are associated pathogenic Clavibacter strains are more frequently en- with the plasmid and/or low GC regions (Additional countered and isolated from the semi-specific medium file 1: Table S2). duringthetomatoseedcertification. The extracellular polysaccharide (EPS). The genomes Toxin-antitoxin system. The presence of the toxin- of all four analyzed Clavibacter strains contained four antitoxin (T-A) genes (YefM Cl_00198, peg.1235 and gene clusters involved in exopolysaccharides production YoeB Cl_00197) in thegenomeofnon-pathogenicClavi- (Additional file 1: Table S9). The EPS production in bacter is intriguing and raises questions concerning their LMG 26808 is expected to effectively occur since all originandpotentialfunctionsinrelationtothephysiology genes involved in that process are functional (no frame- of the bacterium (Additional file 1: Table S2). The YefM shifts, no pseudogenes). LMG 26808 contains almost a
Description: