Chloroplast DNA Phylogeography of Pedicularis resupinata (Scrophulariaceae) in Japan PDF
Preview Chloroplast DNA Phylogeography of Pedicularis resupinata (Scrophulariaceae) in Japan
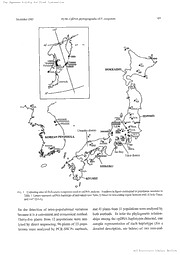
TThhee JJaapapnaesneSeosciee tySociety ffoorPrl anPtlant SSyystsetmaetmicastics ISSN1346-7565 Acta Phytotax .Geobot. 54 (2) 1:63-175 (.2003) ChloroplastDNA Phylogeography Petticularis of resupinata (ScrophulariaicneJaapea)n NORIYUKIFUJII Mtikin oHlerbariu mG,raduate Sbhool ofScie n7cbieg,,o Metropolita nLbeive,:s1i-tiy ,rvfinami-Osaw aH,aehioji, 7bdyo 192-0397 ,lapan Chloropla sDtNA variation in R7dicutar irsesupinata L, (Scrophularia cwaesa est)udied in 48 popula- tions from the Japanes earchipelago and adjacent regions, Ninc distin ccptDNA haplotypes, based on the intergen ispcacer between the trnL (UAA) Stexon and trnF (GAA) ,were recognized. One cpDNA haplotyp ewas distribu wtideeldy (typ Ae), and most ef the other haplotype swere fdund te be geo- graphical lstyructured. Two major clades ( Iand II )were revealed in phylogeneti acnalyses among the haplotype bsy adding the sequence data from the intergen ispcacers of trnT CUGU) and trnL (UAA) S" exon, and aipB and rbcL, The haplotype sof Clade I were widely distribut enodt only within the Japanese archipelago, but also on thc Korean Peninsul aand Sakhalin. Clade II ,howeveg was fbund only in plant sfrom central Honshu and Shikoku ,Japan .These results suggcst that the cpDNA haplotype osf Clade II originated within the Japan and remajn as relics in centra] Henshu and Shikoku .The Japanese origins oftwo endemic taxa, P. resmpinata var. caespitosa Koidz. and var. mictvpltylla Honda arc also discussed. Key words: chloroplast DNA, taxa, haplotypes,Japane saerchipelago, origin, A?dicuiaris resupinata,Scrophulariaceacendemic It has been generally considered that the Japanese groups, There are few studies, however, of the fior awas derive dby invasion sof plant sfrom the phylegenet irecconstruction divergence pattern sof Eurasian or Asian continents via Sakhali nt,he Kuri1 such groups in the Japanes efior ae,xcept for studies islands ,the Korean peninsula, and the Ryukyu of plant sof the Ryukyu Island sCHiramat setu al. island sthroughout past geologica lepochs (Hotta2001, Setoguchi 2001) and on Japanese alpine 1974, Maekawa 1998). As evidences fbr this plant s(Fuj eit ial, 1997, 1999), hypothesi asre the many widespread species in the Pbdicular miessupinata L. (Scrophular iiasceae) Japanes efior athat are also extant on Asian conti- a perennial herb of sunny meadows from the low- nent, It a]so has been reported tha tabout 409/ 6of the lands to the subalpine zone jn Japan .It is distributed plan tspecies in Japan are endemic (Hot t1a974, in eastem Asia in Japan, KDrea, northern to central Murata 1977) ,indicati ntghat relatively many of China ,Manchuria ,Mongolia, Siberia ,Sakhali annd the plant sin Japan originated or differentiat weidth- Kamchatka (Yamaza 1k9i93). The species exhibits in the Japanese archipelago. To understand the considerablc diversi tiyn external morphology and processe isn the evolution of the Japanese fior ai,t is several infraspecif itcaxa have been proposed fbr importan tte clarify the origin ofJapanese endemic plants in Japan .For example, Yamazaki (1981) NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorr PPllanatnt SSyystsetmaetmicastics 164 APG Vbl. 54 recognized fbur varieties: resupinata, oppositijblia1996) ,CpDNA generall ryeflects seed flow and is Miq,, caespitosa Koidz., and micmpdyUa Honda. therefore a good marker for monitoring colonization Yamazaki (199 3re)cognizes fiv evarieties: wsupina- processe s(McCaul e1y994), ta, oppositijb l[iano etxplicitly cited in Flora of In thi sstudM to elucidate the evolutionary pat- Japan] ,mikawana, tet{criijbUa and caespitosa. tems and processe osf infraspeci dfiiycersification, Nlarie mtiycmphJ・'ila Honda is not mentioned and van I irrvestigat etdhe cpDNA variation and phylogeo- resupinata, although appearing in the key ,is not graphi cpattern isn 48 populatio nosf Pladicularis included under the species descripti onA.lthough resupinata from the Japanes eisland asnd adjacent the infraspeci ftirecatment differ asmong taxono- regions. On the basi osfthe results ,I discus tshe ori- mists, varieties caespitosa and micmpitylla are gin ofthe Japanes eendemics, vars. cae.£pitosa and accepted widely (Ohwi 1953, Kitamura et al, 1957, micFz\)hylla, and evaluate Yamazaki' shypothesis. Ylamazak 1i981) ,According to Yamazaki (1981 a)ll, fbur taxa are found in the Japanes earchipelago. Materials and Methods Vlirieti ceasespitosa and micmplp,lla are endemic to Japan ,implyin gthat they originated there .Yama- SZiimpii mnagteriaLy and total DIY4 extraction zaki ( 198 1) inferre tdhe evolutionary histor oyfthe I sampled a tota lof 172 plants from 48 popula- group from observations of their external morphol- tions of Pladieutari sresupinata in the Japanese ogy. He hypothesized that (1 )var. resupinata islands ,the Korean peninsul aS,akhalin, and the entered the Japanese island sfrom the north and Kuril island (sFi g1 ,and fable 1). The samples of that var. caespitosa became differentia ftreodm var. P. yezoensis (sec tPe.dicularis) ,P. schistostagia resupinata in central Honshu; (2 )plant sof var, (sec tB,ieuspidatae), and P. chamissonis (sect. qppositijbli eantered Japan by way of the Korean Orthosiphonia) were also collected to use as out- peninsula and vat micrtzpltylla became diflbrentiategdreups of P, resupinata (sec fte,dicularis) .Iden- from var, oppositijb iiina central Henshu. This tificatio nofthe infraspeci tfaixac ofP. restrpinata is hypothesis has not yet been verified using other mainly according to Yamazaki (1981 ()Tbb l1e) ,I evidence. used the fbllowing morphological characteristics The chloroplast DNA (cpDNA )variation pro- to identif ythe samples: inflorescen ecleongate or vides an opportunity fbr phylogen yreconstruction at shortly spicate or capitate, leave sopposite or alter- the populati olneve l.The geographi sctructuring of nate, and lea fsize. The leave swere drie dand pre- variation may provid einsigh tisnt othe historicalserved in silica gel .Total genomic DNA was biogeography ofspecies (Sol teti asL 1997, Solti &s extracted from O.O lg of dried Ieave susing the Solti 1s998) .Analysi softhe relationships between slightly modified CTAB method of Doyle & gene phylogeny and geographi cdistribut iofotnhe Dickson (1987). phylogenetic greuping swithin a species is termed "intraspecific phylegeography" (Avis ete at, 1987, Outlin eofmethoci dsetfeocrtin qgpDA44 variation Avise 2000) .The chloroplast genome is an attrac- For all plants ,the non-coding region between the tive target for such studies because it is effectively trnL (UAA) 5'exon and trnF (GAA) of cpDNA haploid ,present in multiple copies and does not (Tal)e retl ael.t 1991) was used fbr the recogriition of recombine (Hill i&s Moritz 1990). In most cpDNA haplotype bsy direc stequencing andlor by angiosperms cpDNA is transmitted through the single-strand confbrrnation polymorphism analy- seeds and not through pollen ,although in some sis ofPCR-amplified fragment (sPCR-SSCP mset)h- plan tspecies it is biparent aorl paterna (lMogensenods (Fui eit ial. 1997). The latt emerthod was used NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorr PPllanatnt SSyystsetmaetmicastics Dccember2003 FUJII: CpDNA phylogeography ofP. restv)inata 165 HOKKAID HONSHU .e FIG .1. Collectin sgites of Rsdicular its'est{pinata used in cpDNA analysis. Numbers in figur ecorrespond to populatio nnurnbers in Table 1. Lctter srepresent cpDNA haplobyp eofindividuals {scc 1labl 2e) based on non-coding region between trnL (UAA )5'exon (GAA). and trnF foT the detectio nof intra-populationa lvariation and 41 plant sftom 31 populatio nwesre analyzed by because it is a convenient and economical method, both methods, To infe rthe phylogenet ireclation- Thirty-five plant sflro m12 populations were ana- ships among the cpDNA haplotype dsetected ,one lyzed by direc tsequencing, 96 plants of35 popu- sample representative of each haplotype (fo ra lation swere analyzed by PCR-SSCPs methods, detaile ddescripti osnee, below) of two non-cod- NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorPrl anPtlant SSyystsetmaetmicastics l66 APG Vbi,54 TABr,E 1. Materials a,nd thei rsources, analyzed for cpDNA variation ofAvdicularis resag]inata. PopulatjonNo, Localit yand veucheri No, ofplantsfaXonZ CpDNAhaplotypes5 fledicutar riessttpinata L. (sec Ptl.edicutaris) Russia 1 Northern Kuriles :Onekota nIsland .Alt .1OOm, EZ lhkohashi 28J19 (sAp) 3s44135R5ESRESRESRESREDSDRDDEASERAEASEARAESRESIlliSOPP 2 Centra lKuriles: Simushiv Tsland ,Alt. 20m, fiZ 7inkahashi 28353, 67, 69 (stxp) 3 Centra lKurilcs B:rat Chirpoev Island ,Alt .40m, H, 7lrkahashi .?.84 6(6sAp) 4 Centra ]Kuriles U:rup Island, Alt .1Om, Fl lhkahashi 2S5 73 (sAp) s Centra lSakhalin /25 km west ofPeronaysk, H. lhkahashi 29621 (s,xp) 6 Southern Sakhalin /Pugachevskye (Maguntan )H,. fakahashi 27873 (sAp) 7 Southern Sakhalin: 80 km north ofDolinsk, H. fahahashi 29555 CsAp) 8 Southern Sakha]jn ]20km east ofSokel town, lt 7bhahashi 2910C 22 (s.-p) 9 Southern Sakhalin :2ekm southeast ef' YUzhno-Sakhalinsk J,ldL Sueuki 80502I(TusG) ] 10 Southeni Sakhali n1:0 km east of Korsakov, H. 7inkuhash 2i942J (sA p) 3 11JapanPrimorski j(N440 07' 18.3' 'E,:T350 39' 24.2") ,C. Su.vama17)-4 ,55 (KANA ) 2 12 Northemi Hokkaido: Rebun Island ,A]t .70m, }cA,vg19901O 23221 24 2R3E33SLuOn3ls(52n3o5w4n 234 s3D3s61537s4 13 Eastem Hokl[aido :Mt, Rausu, Rausu, Alt .1300m, kw199005 D 14 Centra lHokl(aido D:aisets uLake. Alt .1OOOm, M4,w199008 OPP A 15 Central Hokkaido/ Mt. Mibari, Alt .1300in ,K}l,N"199009 OPP D 16 Southern Hokkaido: The Erimo Cape, Att .1Om, K]gA・li199006 OPP D 17 Nerthern Honshu: Iwate, Mt, Yakeishj ,Alt .1300m, KAivAi990f2-13 OPP A 18 Northem Honshu: YAmagata, Mt. Gassan, Alt ,1500m, MnK3043I7, 18 OPP Ce 19 Northern Honshu: Yamagata, Mts. Iidc, Alt .2000m, KLi,yd19PO02-3 OPP CAandC 20 Northcrn Honshu: Fukushima, Mt. Nanatsugadakc ,Alt .1500m, ilfrlK30289Z 98 OPP 2t Nerthern Henshu/ Fukushima, Mt Kimen ,Alt .1400m, MwK309687-89 OPP B6 22 Centr aHlonshu, Gunma, Mt. Hakkensan, Alt ,1700m, "tgK304466, 6S, 71 OPP A 23 Centra lHonshu, Nagano. Mt. Amakazari, AIt .1800rn,,Lt4K3224 6784,, 75 OPP H 24 Centra lHonshu, Gunma, Mt. Jizou ,Alt. 1600m, ntAN04324 OPP A 25 Central Eionshu, Nagano, The Ikenotair Maarsh, Alt. 2000m, rmK322471 OPP A6 26 Central Honshu, Nagano, Mt, Chogadake, A]t ,2500m, ,tinK322472, 73 CAE H6A 27 Centra ]Honshu, Nagano, The Kirigamine Highlands ,Alt ,1SOOm, nkK307621, 23 OPP and FCandE 28 Ccntral Honshu, Saitama ,Mt. Futago, Alt .1OOOm,MdK30310S-JO OPP 29 Centra lHonshu, Yirnanashi ,The Ichinose Highlands ,Alt .1300m, ,vfi4K3 1080P-11 OPP AA 30 Ccntra lHonshu. Uimanashi, Mt. Mikuni, AIt .1200m, ,LtdK3041773 OPP and F 31 Centra lHonshu, Yhmanashi, The Suzura nPass, Alt .1400m, nt4K304325, 26 OPP EAandE 32 Centra lHonshu, Yamanashi, Mt, Amar[ ,Alt .1600m, ,LcdK304464-6Z 70 OPP 33 Centr aHlonshu ,Nagano, The Sanpuku Pass ,Alt .2500m, ,w)IE32246P-70 CAE H 34 Centra ]Honshu .Gjfu, The Hokoneko Lake, Alt .800m, ,LdidK304320 MIC E6 35 Centra lHonshu, Aichi ,Mt. Ishinomaki ,Alt. 140rn, iLtgK303502 MfCunknown4 F6 36 Centra lHonshu, Ishikawa, Mts. Hakusan (Ichino Aslct) ,1,500-1700m H 37 Centra lHonshu, lshikawa, Mts. Hakusan (Sannorni Anlet.) 1,8eOm, imK304323 OPP H 38 Western Honshu, IIyogo, Mt Hyonosen, Alt .750m, ,lf.gK328503-08 OPP A 39 Westcrn Honshu, TbttDr iM,t, Daisen, Alt. 800m, ,LcgKj04322 OPP E 40 Shikeku ,Tokushima, Mt. Tsurugi, Alt ,1670m, ,v]K316023, 322933-34 OPP HA 41 Shikoku ,Kochi, The Tengu Highlands ,Alt. 1200-13OOm, ,LmK30323 7-41 epp and J6 42 Kyus}qi ,Kumamo[o, Mt, Ichifus aA,lt .1700m ,,imff303242-45 OPP A NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorPrl anPtlant SSyystsetmaetmicastics December2003 FUJII: CpDNA phylogeography ofP. tesupinata 167 TABLE1. (continued) PopulationNo. Localit yand voucheri N e . of Thxon2 CpDNAhaplotypesS plants NorthKorea 43 P'yong-anbuk-do (N40 056' 14.0' 'E,124U 46' 53.2'' )C,oll .H. Kim, Alt. 600m 3 unknown4 A SouthKerea 4445464K7a4ngg-won-do,Mts.Soraksan(AnasnA)l.t.900-1260m,,LmK322480-83 4545 OPPOPPOPPOPP G6GGA Kang-won-do,Mts.Seraksan(MishiryonAlgt.)8,00-1260m,,it4K322485-88 Kang-won-de, Mt. Odaesan, Alt .800m, ntgN322494, P5 Kyongsangnam-do, Mts. Chirisa nCNegoda nA)lt, .880-1300m, floK322516-20 Kyengsangnam-do,Mts.Chirisa(nChotdacbonAlgt).1,OOO-1400m, 4 OPP A ,LL4K322500-03 Tbtal172plants Outgroups RedicutarisyezoensiMsaxim. (scctfe.dicularis) Centra Hlonshu: Nagano, Mt. Cbegadake, Alt .2SOOrn, ,wAK310467 R schistvstegia Vved, (sec Bti.cuspidota eSteven) Northern Hokl[aido :Rebun Island ,Alt. 20m, L4Acd 198102 R Stcven(secOtr.thosiphoniaH.L.Li) chamissonis Centra lHonshu, Nagano, Mt. Amakazari, Alt ,1900m, whK31046J t2SAP: HekkaidoUniveristyT,USG: TohokuUniversityK,ANA: Kanazawa UniversityM,AK: Makino Herbariurn The intraspeci ftiaxconemy of the P, resupinata were fo11owed by Yhmazaki (198 )1 .The four abbrcviations indicat tehe variety of the species as fo11ows :RES; var, resupinata, OPP; var, qppositijZ) lMiIaC,; var. mieroplrylla, CAE; var. caespitosa. 34sTfhie plant softhe specimen were very young, and we could not identif tyhem. No voucher specimen. The alphabetical characters indicat ethe cpDNA haplotype sobserved in each population s,ee also Fig. 1. The samples were sequenced three cpDNA regions (trn7 :trLn,L-F, and aipB-rbcL> for the present phylogeneti canalyses. ing regions between trnT (UGU) and trnL (UAA) min at 94eC fbr initi adlenaturati ofnb,llowe dby 30 5'exon a,nd aipB and rbeL were furthe srequenced, cycles of denaturati oatn 940C for 1 min, primer annealing at 550C for 1 min and extension at 720C PCR amptipcation and sequencing fbr 2 in{n. The reactions were then extended by 7 Three cpDNA regions were amplified by PCR for min at 720C. nucleotide sequence variation or fbr PCR-SSCPs as The PCR products fbr direc tsequencing were follows t:he non-coding regions between the trnT excised from 1% agarese gels and purifie udsing a (UGU) and trnL (UAA) 3'exon, the trnL (UAA) 5' GENECLEAN II Kit (BIO 101, lnc. )to remove exon and trnF (GAA) ([[laL bete arl.l e19t91), and the non-incorporated primers and nucleotides, aipB and rbcL ([[le r1a9c93h,i Fajii et al, 1997). Sequencing reactions were carried out using an The PCR reaction mixtures contained 50-100 ng ABI Prism BigDye Terminato rCycle Sequencing template DNA, 5pL of 10× PCR buffe r(Takara,Ready Reaction Kit (Appli eBidosystems, Inc.). Japan) ,O.2 mM ef each deoxyribonucleoti 2d.e0, The sequencing reaction products were purified, mM of MgC12, O.4pM ofeach of the primer pairs, concentrated by EtOH precipitation ,and then and 1.0 U ofExTaq DNA polymeras e(Tlak airn aa) applied to an ABI Prism 377 automated DNA total volume of 50pL, The PCR program ran fbr 3 sequencer (Appli eBidosystems ,Inc.) with Long NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorr PPllanatnt SSyystsetmaetmicastics 16S APG Nbi.54 Ranger gels (FMC BioProduct sU,SA). (Erikss eotn aL 1998) .In the MP method, I used a data set includin bgoth substitution data and indel Analysis ofPC:R-ssCRs characters. Indel sin the aligned sequences were PCR-SSCPs of the non-coding region of trnL coded as lfO binar ycharacters in the data matrix, (UAA) 5'exon and trnF (GAA) of cpDNA were Gaps ofmore than 1 bp in lengt hwere treate das detecte bdy fo11owin tghe procedure of Fiiji eit al, being due to single events. All charaeter states, (1997 )T,his region had long sequences (910-930includin igndels ,were specified as unordered and bp) and was digeste bdy restriction enzymes (7i nI,q equally weighted. For the NJ tree, I calculated Takara Co,, Japan )to generat aen adequate lengt hof geneti dcistance bsa,sed solely on substitution data, DNA fragment sfbr SSCP. Gel composition for using Kimura's (1980 t)wo-parameter model in electrophoresis was O,5 × TBE (4 5mM tris-borate, PAUP' 4.0. The bootstra apnalysis was cenducted pH 8,3, 1 mM ED[IA) ,5% glycero alnd O.5 × MDE with 1000 replicates, gel solution (FMC BioProducts )E.lectrophoresis was perfbrmed in O.5×TBE at 250 V for 4.5 h. Results The gel temperature was kept at 1 1 .5 C by use ofa thermostat-controll ewdater circulator. ClpDAC 4hqplompes deteeted [[b recognize the cpDNA haplotype osf,R?dicularis Sequencealignmentandpltylogeneticanalyses resupinata, I determined the sequences ofthe non- Alignment of the sequences was done manually coding region between trnL (UAA) 5'exo nand trnF using the DNASIS-Mac program (Hita cShofitware (GAA) of cpDNA. The nucleotide sequence data Engineerin gJ,apan) .Boundaries ofthe coding and reported in this paper will appear in the DDBJ, non-coding regions were deterrnine dby comparing EMBL and GenBank DNA database usnder acces- the sequences to the corresponding sequences in sion ttumbers AB110781, 110782 ,110828, 110912 IVicotian taabacum L. (Shinoz aekt iaL 1986), to 110930 .The lengt hofthe region varied from Insertionfdele t(iionndse wlerse) general lpylace dso 910-930 bp, and the region include d50 bp of the as to maximize the number ofmatching nucleotides coding region of trnL (UAA) 5'exon and 32 bp of in corresponding sequences. I determined the the coding region oftrnF (GAA) .The polymorphic cpDNA haplotype bsased on the site changes and characters in the region included 43 nucleotide sub- indel osfthe non-coding region between txnL (UAA) stitutions and 17 indel asmong all accessions, and 12 5'exon and trnF (GAA). site changes and eight gaps among the populations Phylogenetic relationships among the cpDNA ofP. nesupinata. In the PCR-SSCP analysis, the haplotypes of P. resupinata were inferre bdy the trnL-F regions of most individua lwesre digested maximum parsimony (MP) and neighborjoining into two or three regions by 7Zi qI, andIobserved (NO methods, using three non-coding regions (tmT fbur ,five o,r six SSCP bands in each lane (Fi g2.). trnL, trnL-trnF, and aipB-rbcL). The most parsi- When a sample exhibited a band pattern of the monious trees were obtained with the PAUP" 4,O same sequence as the known sample, I estimated program (Swoffo 2r0d02) using the braneh and that both samples had the same nucleotide sequence. bound search option. The relative level osf support Through the analysis, I was able to fin idntra-pop- fbr differe ncltades was estimated using bootstrap ulational variation in cpDNA in six populations analysis (Felsens t19e8i5)n based on 1000 repli- (No .20, 27, 28, 30 ,32, and 41, see Tbble 1), cates and decay analysis (Breme 1r988, Donoghue Based on the 12 substitutions and fbur ofthe et al. 1992) using Autodecay version 4.0 programs eight gaps detecte dI ,was able to recognize nine dis- NII-Electronic Library Service TThhee JJaapapnaesneSeosclee tySociety ffoorPrl anPtlSyastnetma tSlcysstematics December2003 FUJII: CpDNA phylogeography ofP. resupinata 169 12 34567 89t..10 ll 12 13 14 l5 16 17 18 , ・'t,i,ll 'e-..{/.l. a n .'.,i"e-g.'e..pl ,.a1 '.i・"'f//ii',:t./.".t."e., ':," v・'. t,- s"u. ',w, t/,.k,S.,t,t:・ges,,,estwiS・'f・・,.,.,,g',//L・i,,,・' li 1.1,geMue,e,w-'J"'it//-ij,・ i・geIS, mefi illllg.-,.}1/i.',tif. 1・"tink,t,.,, ・111,・ /tt ,,t.. .,v,t. a.t , ,"i.tera .wv t ,.i.,.,'.'rte,:it.rl.zl,at{:/.,tsgl, /i/',/l'l:I i1/s l9 ,/f/,-'i -- tw me ,g/w w' Et /tlit/gt/l.' ,-,ss・ ri;.t i,itilsli,・l lllS.l ia・i・isi・i,・e・ei- -・wwt:・meee,.・ i ts-fptetplip t,wi・・ 'sc/,}lil,・ ms ptew '/ . ' 'tttw"1' '' l.I.lee" tees -vS, , ,. 1 t"/d'-e1t"'/ ・,ifX' ''im'aeslxma'ee'siee tt: t.t.ttt ew ' .t ''t. /. tttttt , ・ l/.t:- ・ '・- .・;, '1.,e t・-・ .. /t,' ,-s 'e l'1, ,i'・・e,i,i',i,e,lnil#・ii'l・i・ ,.,,/ ,iigwe-"a'l/d',i'/,ma,,"'}{ t /'{#.- /'-i-/m.li a・:,/ge/{・-filt 2,/・t,w',・"/・・- ''?fi FIG. 2 Example of SSCP band pattern osf cpDNA (non-cod riegniogn between trnL CUAA) 5"exon and trnF (GAA) )ofRidicularis tesupmata ftem fou rpopuiations: 1-5 M;t. Amakazari (popula tNoi.o 2n3, see Tablc 1 or Fig. ]), 6-101 Mt. Chogadake (26 )1,1-15; Ichinos eHighlands (29 )1,6-18; Ikcnotair Maarsh (25). Intra-populatio nvaarliation not shown. tinct cpDNA haplotype isn Rediculari snesupinata tral and westem Honshu (6 ,9, 28, 31, 32, 34, and (Type As-I; Tal)l e2) .For the purpos eofidentifying 39). Haplotypes B and I were each observed in a cpDNA haplotype sand performin gphylogenetic single populatio (n2 1and 4l , respectively). analysjs, fbur indels were omitted. These omis- Redicular iyseioensis ,P. schistostegia and P, sions were located at sequence positio n1s04-106, chamissonis were distinguishe dfrom Haplotype A 142-146 ,555-557 and 864-871 in this region (data ofP. resupinata (sequen cine plant sfrom Mt. not shown), These indels were mostly poly-A or Hyonosen (38) b)y 1O nucleotide substitutions and po]y-T variations, and the mutual homology of their six indel sn,ine site changes and eight gaps, and sites was ambiguous, Haplotyp eA was fbund to be 25 nucleotide substitutions and fiv cindel sr,espec- distribute wdidely througheut the Japanese islands,tively. the Korean peninsula ,northeastern Russia, and Sakhali (nFi g1.) .Most ofthe remaining haplotypes Pdylogenetie analysis among opDACt lhaptompes were geographicall ylocalize dH.aplotyp eH was To clarify the phylogenetic relationship among the found in the highland sof central Honshu (23 2,6, nine-cpDNA haplotypes of Pedicularis resupina- 33, 36, and 37) and on Mt, Tsurugi in Shikoku ta,l furthe srequenced two non-coding rcgions of (40 )I. recognized haplotyp eC in four populations cpDNA representative of each haplotype of this from northern Honshu (18 ,19, 20, and 28), and species and three outgroup species (Tab l1)e ,[Irhe haplotyp eF in the three populations fi:o mcentral lengt hofthe intergen riegcions between trnT (UGU) Honshu (27 3,0, 35) .Most populatio nons the Kuril and trnL (UAA) 5' exon varied from 713-715 bp, islands and in Hokkaido, Japan, possessed the and the region include d10 bp ofthc coding region cpDNA ofHaplotype D (1- 41,2, 13, 15, and 16) .In oftrnZ In this region, 57 nucleotide substitutions Korea ,the three populatio nfsrom the central penin- and fiv eindel wsere found among all accessions (12 sula exhibited an endemic haplotyp e(ty pGe) (44-sequences), and 17 site changes and thrce indels 46). Haplotype E showed a discontinuo udsistrib-were detected within the cpDNA haplotype osfP. ution patter ni;t was fbund on Sakhalin and in cen- resupinata. The lengt hof the intergen irecgions NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorr PPllanatnt SSyystsetmaetmicastics 170 APG Xlo1 .54 TABLE2.ThechloroplastDNAhaplotypesofAedicularisresupinata. trnL intron trnL-Fspacer Position 116 139 126471 397 656 667 7888 99Ol71 ggo soe4 Hapletypc 5S6 8092 ABCDEFGHI Att-ttTitkcGtGt*tcC cde Tde ATktde TGt'-'T,kdtetCcGkdtetdeAGtTtddeekAtATdetdet*GGG ±±±tttdetsikttde t±tk-de+dekt-kde tttkkdet* trkRdekdeTT ± ± dekde t"-t-dett ± dek+tkttt-tde ± k tttttt deik ±± ± tt-tkdettt t'tTT kGt k-tk" dedede iktttAtAGAG ±dektttttktttAACAAAAACAAA ttG tttttkkde ± c de t ttttktt* ± AAACttsde *tt t tttt"t de k - [VCTTATCA ± A dot denote ssite changes or indel sidentie atlo the characteristics of Haplotype A. between aipB and rhcL varied from 732-739 bp, and 74% bootstra pprobabilit yagainst the outgroups. 15 site changes and seven gaps were detect aemodng TWo major clades were revealed within the haplo- all accessions, and five site changes and three indels types of P. resupinata (Clade sanId II) ,Clade II were detected in the intraspecifi clevel of P, comprised cpDNA hqplotypes H and I with a boot- resupinata. The total length of the combined data strtip value of 100%. Clade I include adll remaining set after multiple alignmerrts of the three regions was haplotypes (A-G )with a 1O09/ b6ootstra pprobabil- 2473 bp, while 117 site changes and 30 gaps were ity .Within thi sclade, Haplotype G was sister to the inferre damong all accessions; 34 site changes and other haplotype (sC]ad IeII ;A-F) with a bootstrap 14 indel swere detecte dat the intraspeci fleivcel. value of 1009,6 .The phylogeneti crelationships The DNA divergenc evalues (uncorre cp-tdeids- within Clade III were unresolved polytomies, but tances, Kumar et aL 1 993), excluding indel asmong haplotypes E and F formedaclade (Clad IeV) with the cpDNA haplotypes, ranged between 0 and a bootstra vpalue of 88%, 1,11%. In the MP analysis, two ofthe five gaps in The NJ tree based on the Kimura two-para- the trn7:L region and four of the eight indels of meter model is shown in Fig, 4. The topelegy ofthe the aipB-rbcL region were not used (dat naot NJ tree was almost the same as tha tofthe MP tree. shown), The monophyly ofthe haplotype osfP. resupinata, Wagner parsimony analysis of nine cpDNA however ,was weakly supported (38%), haplotype osfRedicularis resupinata based on the characteristics ofboth site changes and indel rsesult- Discussion ed in 105 parsimoniou tsrees using the branch and bound search option. The trees required 153 steps; TWo major clades (Clad Ie asnd II )were revealed in Consistenc ylndex including uninformative char- the cpDNA haplotype sof Pledicular irsesupinata acters (CI =) O,9281 ,Retentio Inndex (RI -) O,8981. with high bootstra pprobabilit y(Fig s3 .and 4), The strict consensus tree (MP tree) ofthe 105 trees suggesting that the haplotypes of each clade is shown in Fig .3, The MP tree displaye tdhe fo1- diverge dfrom a common ancestral genopae .Clade lowingphylogene treilcationships. Firs tt,he cpDNA I included seven haplotypes a,nd Ciade II com- haplotype sofP. resupinata formed a clade with prise donly two haprotype s(typ eHs and I) .The NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorr PPllanatnt SSyystsetmaetmicastics December 2003 FUJIIiCpDNA phylogeography ofP. resupinata 171 haplotype osfthe latte crlade wcre distribut oendly by having capitate infiorescence (sKoidzu m19i25, in central Honshu and Shikoku, Japan (Fi g1.) in Kitamura 1958, Yhmazaki 1981, 1993, Shirr}izu populatiens at relatively high altitudes (Tab l1e), 1982). Although plants in other populatio nwsith The remaining haplotype sb,elongin gto Clade I, the cpDNA pattem ofClade II (23 3,6, 37, 40, and were distribut ewiddely in the Japanes eislands 41) were identifi aesd var. oppositijbli av,ar. cae- (excludi nShgikoku), the Korean peninsula, spitosa was observed only in Clade II .Variety eae- Sakhalin a,nd the Kuril islands .In short, the hap- spitosa may have differentia ftreodm the common lotype sof Clade II have a restricted distributionancestor of Clade II. and are endemic haplotype isn Japan. These find- In Clade I, the cpDNA haplotyp Ge was sister ings may indica ttheat the cpDNA haplotype sof to haplotypc As-F (Clad IeII )with high bootstrap Clade II originated within the Japanes eislands, probabilit y(Fig s3 .and 4) ,Haplotype G was ob- where they remain as relics in central Honshu and served only in central Korea ;the nonhern and south- on Shikoku. ern Kerean pepulation sshowed the cpDNA oftype In Japanes ealpine plants t,he endemic clade A in Clade III (Fig .I) . This evidence suggests distribu tien dcentral Honshu was inferr efdrom the that Haplotyp eG diverg eeadrly from the common phylogeographical analysis using cpDNA; ancestor of Clade I and remained in the Sorak Pediculari schamissonis Steven (Foj eit ial. 1997), mountain area of the Korean Peninsula .In my Primula cuneijblia Ledeb, (Foj eti ail, 1995, 1999). fiel odbservations in 2000 in the Sorak mountains, I Both species occur in the subalpine-alpine area of saw only plant swith a white corolla. They were Japan, and are distribu tien dthe Nonh Pacifi ccoasta1 describe ads Rrdicular iressupinata var, resupinata area from the Japanese archipelago nonheastwards f. atbWora Y, N, Lee (Le e1996) .Most of the to southwest Alaska. These studies showed that plant osfP. nesupinata have a pinkis hred or reddish one clade was distribut eidn central Honshu and purpl ecorolla. I did not see any pinkish flower dsm- the other clades were present in northern Honshu ing my visit, which may indica tteha tmorphologicat and northward. They assumed that the ciade dis- differentia tait olne,as itn corolla colog has occurred tribute din central Honshu entered the Japancse in the populatio nofs the Sorak Mountains in Korea. island searlier than did the northern clades, and The chloroplast genomes of hapletyp eAs-F of considered that central Honshu played the role of a Clade I!I had the most derivati vpeosition in the- refugia fbr the species in the interglac ipearliods of trees, but the geneti cdifferentiatio nasmong the the Pleistoce neepoch. In the prcsen tanalyses, hapletype swere relatively small (Fig s3 .and 4). clade II of P, resupinata is endemic to central The distribut iaornea of the clade was wide, espe- Honshu and Shikoku (Fi g1.) ,although the distrib- cia]ly that of Haplotype A (Fi g1.). These find- ution patter nof the clade diffe rfsrom those ofP, ings suggest that the genomes ofClade III diverged chamissonis and Pr. euneijblia. Cladc II of P. and cxpanded their distribut iaroena more recently resupinata may have arisen durin gthe migration than those of types G-I in northeastern Asia. processes that took place durin gchanges in the cli- Furthermor eth,ey may have different iian ctenetdral matic during the Plejstocene. Honshu (typ eB sand F), northern Honshu (type Two population fsrom central Honshu (nos.C), and in the Kuril island sand Hokkaido (type 26 and 33) ,identifi aesd Pedicular inesstrpin'ata var, D). caespitosa, possessed the cpDNA profi lofeClade II Haplotype sE and F of Clade III fbrm their (ty pH)e. [[Ih ivsariety, which occurs in the subalpine own clade (Cla dIeV) ,Most plant sofClqde IV are area ofcentral to northern Honshu, is distinguished in central Honshu, but some are also on Sakha]in NII-Electronic Library Service TThhee JJaapapnaesneSeosciee tySociety ffoorr PPllanatnt SSyystsetmaetmicastics 172 Al)G Xlo1 .54 PR type A PR type B PR type C I PRtypeD I PRtypeE IV PRtypeF PRtypeG PR type H II PRtypeI Pedicularisyezoensi,s P. schistostegia P.chamissonis F[c i3.. Stri ccotnsensus of 1OS most parsimDnlou strees for R]dicuiar irsestu)inata cpDNA haplotype sdcrii, efdrom trn 7LtrnL ,trnL- F and aipB-rbcL non-coding rcgions, Nunibers above branche sare bootstr vaaplues, in percentage sb,ased on 1,OOO replicates, and decay indexes ,rcspectively, So]id and open bars represent sitc changes and indels r,espectively. Bar with asterisk indicates homoplasti ccharacters. Numbers on branches are nurnbers ofsite changes and indels ,respectively. (Fi g1,) .Individua lisn two populatio nfsi/o mthe lia ,and vars. resupinata and caespitosa entered Ibkai distri (cntos 3.4 and 35) with cpDNA of clade Japan from the north. Ifhis hypothes iiss correct, it IV were identifi easd Pedicularis resupinata var, is expected that individua lofs vars. micropIrylla micmplp?lta. Variety microphylla occurs mainly and qppositijbli awould fbrm a clade, and vars. in the [Ibka idistri coft Honshu and is among the resupinata and cae,!pitosa would constitute a sister "Tokai hjll yland elements," which are relic taxa in group, In my cpDNA analysis, however ,plants the region surrounding Ise Bay (Ued a1989, 1994), identifie das var. oppositijbli aoccurred in all of It is believe dthat taxa of this association have the clades (I-I Vw)hi,le those identifie das vars. adapted to the unique, small, peat-fre eswamps and resupinata and caespitosa were placed in different marshes in the region, In this analysis, var, micro- clades ( Iand II )(Figs a.n3d 4) .Yamazaki' hsypoth- pbylla was recognized only in Clade IY suggesting esis was therefbre not supported by cpDNA analy- that plants ofvar. micmplp,lla differentia ftreodm a sis. CpDNA capture through introgressive part of clade. hybridizati oorn lineag esorting, which has been According to YamazakVs (1981 h)ypothesis reported in many plan ttaxa by Wendel & Doyle on the evolutionary histor oyfPladicularis resupina- (1998 )could explain the discrepanc y,To verify ta ,var. micmpfp]ila is derive dfrom var, oppositij2)-the possibil iwtey ,need to examine the phylogenetic NII-Electronic Library Service
