Bacterial diversity of autotrophic enriched cultures from remote, glacial Antarctic, Alpine and ... PDF
Preview Bacterial diversity of autotrophic enriched cultures from remote, glacial Antarctic, Alpine and ...
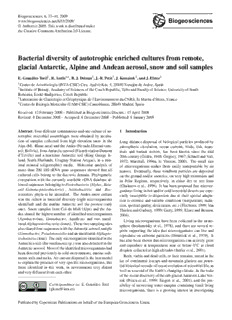
Biogeosciences,6,33–44,2009 Biogeosciences www.biogeosciences.net/6/33/2009/ ©Author(s)2009. Thisworkisdistributedunder theCreativeCommonsAttribution3.0License. Bacterial diversity of autotrophic enriched cultures from remote, glacial Antarctic, Alpine and Andean aerosol, snow and soil samples E.Gonza´lez-Toril1,R.Amils1,4,R.J.Delmas3,J.-R.Petit3,J.Koma´rek2,andJ.Elster2 1CentrodeAstrobiolog´ıa(INTA-CSIC)Ctra. AjalvirKm. 5,28850Torrejo´ndeArdoz,Spain 2InstituteofBotany,AcademyofSciencesoftheCzechRepublic,TeboandFacultyofScience,UniversityofSouth Bohemia,E`eske´ Bud`ıjovice,CzechRepublic 3LaboratoiredeGlaciologieetGe´ophysiquedel’EnvironnementduCNRS,StMartind’He`res,France 4CentrodeBiolog´ıaMolecular(UAM-CSIC)Cantoblanco,28049Madrid,Spain Received: 12February2008–PublishedinBiogeosciencesDiscuss.: 15April2008 Revised: 8December2008–Accepted: 8December2008–Published: 8January2009 Abstract. Fourdifferentcommunitiesandonecultureofau- 1 Introduction totrophic microbial assemblages were obtained by incuba- tion of samples collected from high elevation snow in the Long distance dispersal of biological particles produced by Alps(Mt. Blancarea)andtheAndes(NevadoIllimanisum- atmospheric circulation, ocean currents, birds, fish, mam- mit,Bolivia),fromAntarcticaerosol(FrenchstationDumont mals and human vectors, has been known since the mid d’Urville) and a maritime Antarctic soil (King George Is- 20thcentury(Gisle´n,1948;Gregory,1967;SchnellandVali, land, South Shetlands, Uruguay Station Artigas), in a min- 1972; Marshall, 1996a, b; Vincent, 2000). The small size imal mineral (oligotrophic) media. Molecular analysis of of microorganisms makes them easily transportable by air more than 200 16S rRNA gene sequences showed that all masses. Eventually, these windborn particles are deposited cultured cells belong to the Bacteria domain. Phylogenetic onthegroundand/orsnow/ice, onveryhighmountainsand comparison with the currently available rDNA database al- in Polar Regions, respectively, in either dry or wet form lowedsequencesbelongingtoProteobacteria(Alpha-,Beta- (Chalmers et al., 1996). It has been proposed that microor- and Gamma-proteobacteria) , Actinobacteria and Bac- ganismslivinginhotand/orcoldterrestrialdesertsareespe- teroidetes phyla to be identified. The Andes snow culture cially susceptible to dispersion due to their special adapta- was the richest in bacterial diversity (eight microorganisms tion to extreme and variable conditions (temperature, radia- identified) and the marine Antarctic soil the poorest (only tion,spectralquality,desiccation,etc.)(Flechtner,1999;Van one). Snow samples from Col du Midi (Alps) and the An- ThielenandGarbary,1999;Garty,1999;ElsterandBenson, dessharedthehighestnumberofidentifiedmicroorganisms 2004). (Agrobacterium, Limnobacter, Aquiflexus and two uncul- Living microorganisms have been collected in the strato- turedAlphaproteobacteriaclones).Thesetwosamplingsites sphere (Imshenetsky et al., 1978), and there are several re- alsosharedfoursequenceswiththeAntarcticaerosolsample ports supporting the idea that microorganisms can live and (Limnobacter,PseudonocardiaandanunculturedAlphapro- reproduce on airborne particles (Dimmick et al., 1979). It teobacteriaclone). Theonlymicroorganismidentifiedinthe hasalsobeenshownthatmicroorganismscanactivelygrow Antarcticasoil(Brevundimonassp.) wasalsodetectedinthe and reproduce at temperatures near or below 0◦C in cloud Antarcticaerosol.Mostoftheidentifiedmicroorganismshad dropletscollectedathighaltitudes(Sattleretal.,2001). beendetectedpreviouslyincoldenvironments,marinesedi- Both,viableanddeadcells,ortheirremains,storedinthe mentssoilsandrocks. Aircurrentdispersalisthebestmodel ice of continental icecaps and mountain glaciers are poten- toexplainthepresenceofveryspecificmicroorganisms,like tialhistoricalrecordsofrecentevolutionofmicrobiallife,as those identified in this work, in environments very distant wellasarecordoftheEarth’schangingclimate. Inthewake andverydifferentfromeachother. oftherecentdiscoveryofthesub-glacialAntarcticLakeVos- tok (Priscu et al., 1999; Siegert et al., 2001), and the pos- Correspondenceto: E.Gonza´lez-Toril sibility of recovering water samples containing fossil living ([email protected]) microorganisms, there is a growing interest in investigating PublishedbyCopernicusPublicationsonbehalfoftheEuropeanGeosciencesUnion. 34 E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil thetransportoflivingorganismsviaairoverlargedistances, withhighcontentofbacteriaproducingpigmentedcolonies e.g.coldglacialregions. werechosenforthisstudy. Recently,thecompositionofmicro-autotrophs(cyanobac- Intheyear2000,twosnowpitsweredugatveryhighel- teria and algae), micro-fungi (hyphae and spores), bacteria evation sites in the Mt. Blanc area of the Alps: one (0.6m (rod,cocciandpigmentedbacteria),yeastandplantpollenin deep) at Col du Midi (elev. 3532ma.s.l., 17 May) and the remoteaerosol,depositedsnow,andicehavebeenevaluated other(2mdeep)atColduDome(elev.4250ma.s.l.,31Au- (Elster et al., 2007). As a product of this study cultivable gust) for European sampling. Snow collected from recent autotrophic pigmented microorganisms were obtained from deposits contained several visible dust layers. This dust is alpine snow (Alps and Andes) and aerosol (Antarctic) sam- knowntobetransportedairbornetotheAlpsfromtheSahara ples. desert(Oeschgeretal.,1977;Wagenbach,1989;Angelisand Ithasbeensuggested(Imshsnetskyetal.,1978,Christner Gaudichet,1991). Thedustlayerscorrespondtoshortevents etal., 2000)thatthepresenceofhighlypigmentedbacterial (outbursts) that occur under meteorological conditions such coloniesinthemesosphereandinglacialicewasassociated as a low descend over the western North Africa (Morocco) withtheneedtoprotectcellsfromharmfulUV-radiationdur- producing a southern wind over Italy and southern France. ingatmospherictransportandexposureonthesurfaceofthe The time corresponds to a few hours up to some days. At glaciers. Morphologically similar bacterial specimens have Col du Dome the accumulation rate is variable, but from a also been observed in soils in the Arctic Svalbard (unpub- pit,theageofthelayermaybeafewmonthsuptoayear. lisheddata,Øehakova´ etal.,2009). Theseobservationssup- IntheAndes,surfacesnowblockswerecollectedin2000 porttheideathatthistypeofmicroorganismscommonlyde- at the summit of Nevado Illimani, Bolivia (16◦370S, velopincolddesertecosystemsandareeasilytransportedvia 67◦460W,CordilleraReal, elevation6350m). TheBolivian aerosoltoboth,shortandlongdistances. AndesaresurroundedbytheAltiplano,ahighaltitudedesert (mean elevation: 3700ma.s.l., Clapperton, 1993). This re- In this work we report the identification, using molecu- gion, in particular, contains large salt flats called “salares” lar ecology methodologies, of autotrophic microorganisms (Risacher, 1992), which are an important source of dust for abletogrowoligotrophicallyinenrichmentculturesofsam- the regional atmosphere during the dry season. From size plesobtainedfromalpinesnow(AlpsandAndes)andaerosol and depth of snow-dust layer it has been estimated that the (Antarctic). Forcomparisonthecultureofpigmentedbacte- age of the collected sample from this site was the same or riaisolatedfrommarineAntarcticsoil(KingGeorgeIsland, similaroftheMt.Blancarea. SouthShetland)wasalsoanalysed. Samples from the Alps (Col du Midi 6 samples+blank, Col du Dome, 9 samples + blank) and the Andes (2 sam- ples)wereanalysed(Elsteretal.,2007). Eachsnowsample 2 Materialandmethods containedabout0.5to1kgofsnow.Thesurfacelayer(about 3–5cm)fromthesnowsampleswascutoutandonlythecen- 2.1 Sitedescriptionandsamplecollection tralpartofthesnowblocksweretakenforanalysis(formore detailsonsamplepreparationseeElsteretal.,2007). Inthis Theaerosolsamplesoriginatedfromasetoffilterscollected set 88% of analysed samples contained bacteria producing between1994and2000atthecoastalAntarcticStationDu- pigmented colonies (Elster et al., 2007). From these sets, mont d’Urville (66◦400S, 140◦010E) for long-term atmo- the samples with the highest content of pigmented bacteria sphericchemistrystudies. Thisstationissituatedonasmall were concentrated by lyophilisation (two Alps and one An- island, a few hundred meters offshore from the mainland. dessamples)forfurtheranalysis. Local meteorological conditions were described by Pe´riard In the austral summer season of 2005, soil samples were & Pettre´ (1992) and Ko¨nig-Langlo et al. (1998). The main collectedfromthevicinityoftheUruguayAntarcticStation features of the chemical composition of the aerosol can be Artigas, King George Island, South Shetlands. All sam- foundinWagenbachetal.(1998),Minikinetal.(1998),and ples were collected using pre-cleaned glass vials (aerosol) Legrandetal.(1998).Alltogether,13Antarcticaerosolsam- and sealed plastic bags (snow and soil), respectively, trans- plesandcontrolswerecollectedonGelmanZefluor®filters ported frozen to the laboratory in Grenoble and/or in Tebo (47mm diameter, 0.5µm pore size) by drawing in air at a andstoredfrozenuntilfurtheranalysis(formoredetailssee flow rate of 1.5m3h−1. The sampling interval was 20 h in Elsteretal.,2007). summer(fromNovembertoFebruary)and40htherestofthe year,whichcorrespondsto30and60m3 ofair,respectively. 2.2 Samplepreparationandcultivation Alldevicesusedwerewashedthreetimeswith18.2M(cid:127)cm MilliQ® water, except for the filters. Collected filters were Tominimizepossiblecontaminations,allpost-samplingma- kept under cool, dark and dry conditions (−25◦C). In 38% nipulations were done in a UV-sterilized laminar flow hood of analysed aerosol samples the bacteria that produced pig- using sterile glass vials. To retrieve samples from the snow mentedcolonieswererecorded(Elsteretal.,2007). Samples pits an upper layer of about 2cm was sliced with a sharp Biogeosciences,6,33–44,2009 www.biogeosciences.net/6/33/2009/ E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil 35 knife. The samples were dried off at a temperature of 359and805forCyanobacteriaphylum(Nu¨beletal.,1997) −40◦C in a special sterile lyophilization device (Lyovac wereperformed.ThesegeneswereamplifiedbyPCRinmix- GT2,Leybold-Heraeus,Germany). Glassvesselscontaining turescontaining20–30ngofDNAper50µlreactionvolume, the solid deposits were rinsed with 10–15ml of re-distilled 1×PCRbuffer(PromegaBiotechIberica,Spain),2.5µMof water. eachofthedeoxynucleotides(AmershamBiosciences,UK), Aerosolfilterswerecutintoquartersandonequarterwas 2.5mMMgCl2, 1mgmL−1 bovine serum albumin (BSA), used for enrichment cultures. Each snow and aerosol sam- 500mMofeachforwardandreverseprimersand0.025U/µl ple was cultured in a sterile glass bottle (25ml in volume) of Taq DNA polymerase (Promega Biotech Iberica, Spain) inBG-11culturemedia(asdescribedinBischoffandBold, (Table 1). PCR conditions were as follows: initial denatu- 1963). Eachglassbottlewasfilledwithabout10mlofster- ration at 94◦C for 5 min, followed by 30 cycles of denatu- ile medium (for more details see Elster et al., 2007). Sus- rationat94◦Cfor40s, annealingat52◦Cfor1minforthe pensionsof10gsoil+90mlsterilewaterwerehomogenized Bacteria domain, 56◦C for the Archaea domain, and 60◦C usingultrasonicationfor4minforsoilsamplesanalysis. For for the Cyanobacteria phylum. Positive and negative con- enrichment cultures 1ml of lyophilized snow or solid sus- trolswerealwaysusedineveryPCR.Large16SrRNAgene pensionsoraquarterofafilterwereused. Forcolonyisola- fragments (>1400bp) were purified by GeneClean Turbo tion0.1mlofenrichmentcultureswerespreadonPetridishes Column (Q-Bio Gene Inc., CA, USA) and cloned using the containing1.5%BG11agar(Elsteretal.,1999). Fourrepli- TopoTaCloningKit(Invitrogen,CA,USA).Clonedinserts cateswereperformedforeachagarculture.Glassbottlesand were amplified using PCR conditions described above and Petridisheswerecultivatedinanilluminated(∼100W/cm2) weredirectlysequencedwithaBig-Dyesequencingkit(Ap- refrigerator(temperature5–8◦C)withalightregimeof18h plied Biosystem) following the manufacturer’s instructions oflight, 2hofUV-Bradiation(germicidelamp)and4hof (Gonza´lez-Toriletal.,2006). darkness. Thegermicidelampwasusedtosterilizethecul- ture growth area (UV-B light did not penetrate through the 2.5 Clonelibraryanalysis glass bottles). Experimental bottles were shaken every 2– 3 days. After 1 and 2 months of cultivation, the contents Sequences were analyzed using BLAST at the NCBI of bottles and Petri dishes were analyzed under the light database (http://ncbi.nlm.nih.gov/BLAST) and added to the microscope (Olympus BX 60). Aliquots of samples were mostimportantBLASThits,toreachanalignmentof50000 analyzed by fluorescence after staining with the DAPI flu- homologous bacterial 16S rRNA primary structures by us- orochrome (EFM Olympus BX 60) (Zachleder and Cepa´k, ing the ARB software package aligning tool (http://www. 1987)andtransmissionelectronmicroscopy(Jeolequipment arb-home.de) (Ludwig et al., 2004). The rRNA alignments JEM1010). were corrected manually and alignment uncertainties were Fiveoligotrophicenrichmentcultures,fromeachsampling omitted in the phylogenetic analysis. Phylogenetic trees station(AntarticStationDumontdU´rville;ColduMidi,Col were generated using parsimony, neighbour-joining, and du Dome, Nevado Illimani and Uruguay Antarctic Station maximum-likelihood analyses with a subset of 200 nearly Artigas)wereselectedforphylogeneticanalysis. full-length sequences (>1400bp). Filters, which excluded highly variable positions, were used. In all cases, general 2.3 DNAextraction tree topology and clusters were stable and a consensus tree was generated (Gonza´lez-Toril et al., 2006). Sequences ob- One ml of each enrichment culture was used for DNA ex- tained in this study have been deposited in the EMBL se- traction. Fast DNA Spin kit for soil (Q-Bio Gene Inc., CA, quence database under accession numbers from EU429484 USA)wasusedaccordingtothemanufacturer’sinstructions. toEU429508. Todisruptthecells,themixtureofceramicandsilicabeads providedinthekitandthreepulsesof40satspeed5.5ofthe FastPrep bead-beating instrument (Bio 101) were applied. 3 Resultsanddiscussion Aftertheextraction,DNAwaspurifiedbypassagethrougha GeneCleanTurbocolumn(Q-BioGeneInc.,CA,USA)and Followingtheprotocolsforenrichmentculturesofphotoau- quantifiedbyethidiumbromide-UVdetectiononanagarose totrophicmicroorganismspresentinsnowsamplesfromthe gel(Gonza´lez-Toriletal.,2006). Alps and the Andes and in an aerosol sample from the Antarctica,pigmentednon-photosyntheticprokaryotesgrew 2.4 16SribosomalRNAclonelibraryconstruction successfully in the extremely poor nutritional conditions of theselectedmedia(Fig.1)(Elsteretal.,2007). Similarpig- PCRamplificationof16SrRNAgenefragmentsbetweenE. mented bacteria have also been observed in soils in King colipositions8and1507forBacteriadomain(Lane,1991), George Island, South Shetland Island group as well in the between E. coli position 25 and 1492 for Archaea domain Arctic Svalbard Islands (øehakova´ et al., 2008). It appears (AchenbachandWoese,1995),andbetweenE.coliposition that this type of bacteria commonly develop in cold desert www.biogeosciences.net/6/33/2009/ Biogeosciences,6,33–44,2009 36 E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil Table1.PrimersusedforPCRamplification Primera Targetsiteb Sequence(5’to3’) Specificity Reference 8F 8–23 AGAGTTTGATCMTGGC BacteriaDomain Lane,1991 25F 9–25 CYGGTTGATCCTGCCRG ArchaeaDomain AchenbachandWoese,1995 1492R 1492–1513 TACGGYTACCTTGTTACGACTT Universal AchenbachandWoese,1995 Cya359F 359–378 GGGGAATYTTCCGCAATGGG Cyanobacteriaandclhoroplasts Nu¨beletal.,1997 Cya106F 106–127 CGGACGGGTGAGTAACGCGTGA Cyanobacteriaandclhoroplasts Nu¨beletal.,1997 Cya781R(a)c 781–805 GACTACTGGGGTATCTAATCCCATT Cyanobacteriaandclhoroplasts Nu¨beletal.,1997 Cya781R(b)c 781–805 GACTACAGGGGTATCTAATCCCTTT Cyanobacteriaandclhoroplasts Nu¨beletal.,1997 a F(foward)andR(reverse)indicatetheorientationsoftheprimersinrelationtotherRNA.b PositionsaregivenaccordingtotheE. coli numberingofBrosiusetal.,1981.cReverseprimerCYA781RisanequimolarmixtureofCYA781R(a)andCYA781R(b). any of the DNA extracted from the five enriched cultures. Theseresultsagreewiththemicroscopyanalysisofthecul- tures (Elster et al., 2007). Also, no Archaea were detected byamplification. PCRamplificationof16SrRNAgenesus- inguniversalbacterialprimers, followedbycloningandse- quencingproducedaround200sequences,whichwereused to identify the different bacteria present in the enrichment cultures by comparison with the NCBI database. The re- trieved sequences were added to a database of over 50000 prokaryotic 16S rRNA gene sequences using the aligning tooloftheARBsoftwarepackage(http://www.arb-home.de) (Ludwig et al, 2004). Every sample was processed in the same way: positive PCR products were cloned and 50 pos- itive colonies were chosen and sequenced. Around 50 se- quences were obtained for every enrichment culture. All of them were aligned using the ARB software. With the sequences aligned we built a distances matrix that allowed Fig.1. Displayofpigmentedmicroorganismsenrichedfromsnow different OTUs (operational taxonomic units) to be identi- andaerosolsamples. fied: 3 for Col du Dome, 7 for Col du Midi, 8 for Illi- mani, 1 for Artigas and 6 for aerosol (Antarctis). One rep- resentativesequencefromeveryOTUwasselectedforphy- ecosystems and is easily transported with aerosols at both, logenetic analysis. With these selected sequences phyloge- shortandlongdistances. In1978,Imshsnetskyetal. hadal- netictreesweregeneratedusingparsimony,neighbourjoin- readystudiedpigmentationofviablebacteriarecoveredfrom ing,andmaximum-likelihoodanalyseswithasubsetof200 the mesosphere. These authors suggested that the pigments nearly full-length sequences (>1.400bp). Filters excluding produced by these bacteria are associated with the need to highlyvariablepositionswereused. Inallcasesgeneraltree absorbharmfulUV-radiation. RecentlyDuetal.(2006)and topology and clusters were stable. Thus, consensus trees Mayilrajetal.(2006)havedescribedtheoccurrenceofpig- weregenerated(Figs.2–6). mented bacteria from various marine and soil ecosystems, respectively. After the analysis of the generated sequences, represen- Due to the characteristics of the cultures and the origin tatives of three bacterial phyla: Proteobacteria (Alpha-, of the samples it was considered of interest to identify the Beta-andGamma-proteobacteria),ActinobacteriaandBac- microorganismspresentinthedifferentcultures,toevaluate teroidetes, were identified (Fig. 2). Seven close relative the diversity corresponding to this very specific type of mi- microorganisms were identified for the snow sample from croorganismsandeventuallytocomparetheresultsobtained Col du Midi (Alps). Three sequences corresponded to in geographically dispersed sampling sites. A commercial the Alphaproteobacteria: one as a possible member of soil DNA extraction kit was used to extract DNA from the the genus Agrobacterium (closest relative: clone IrT-J614- enrichment cultures from particulate matter present in high 14, retrieved from an environmental sample of a uranium elevation snow in the Alps and the Andes, from an Antarc- mine (Selenska-Pobell, 2002) (Fig. 3), the other two; close ticaerosolandamaritimeAntarcticsoil. Nooxygenicpho- to clones B3NR69D12 and AP-12, detected in an under- totrophic Bacteria were amplified using specific primers in ground cave (Northup et al., 2003) and in a marine estuary Biogeosciences,6,33–44,2009 www.biogeosciences.net/6/33/2009/ E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil 37 Fig.2.Prokaryoticphylogenetictreeshowingthephylainwhichenrichedmicroorganismsfromsnow,aerosolandsoilhavebeenidentified. (NCBI accession number AY145551, unpublished) respec- Eightcloselyrelatedmicroorganismswereidentifiedfrom tively. TwosequencescorrespondedtotheBetaproteobacte- the Andean snow sample (Nevado Illimani). Three se- ria: oneapossiblememberofthespeciesHydrogenophaga quencescorrespondedtotheAlphaproteobacteriaclass: two palleronii (Fig. 4), identified by FISH in lake snow aggre- related to the Agrobacterium (clone IrT-J614-14) (Fig. 3) gates (Schweitzer et al., 2001), and the other close to a and the clone B3NR69D12, both identified in Col du Midi; clone of the genus Limnobacter, D-15, isolated from un- andonerelatedtotheunculturedEV818CFSSAHH29clone, derground mineral water (Loy et al., 2005). One sequence which was identified from subsurface water in the Kalahari correspondingtotheGammaproteobacteriaexhibitedahigh Shield, South Africa (NCBI accession number DQ336984, homologywithPseudomonaspseudoalkaligenes,whichwas unpublished). Two sequences corresponded to the Betapro- isolated from a marine sediment (NCBI accession number teobacteria class: Hydrogenophaga palleronii (Fig. 4) and AF286035,unpublished). Onesequencecorrespondedtothe LimnobactercloneD-15,bothalsoidentifiedinColduMidi. phylum Bacteroidetes, with high homology with Aquiflexus One sequence exhibiting high homology with Aquiflexus balticus(Fig.5),alsoretrievedfromamarinesediment(Bret- balticus(Fig.5)ofthephylumBacteroidetes,previouslyde- taretal.,2004). scribedinColduMidi,wasretrievedfromtheAndesenrich- The other sample from the Alpes (Col du Dome) exhib- ment culture. The last two sequences corresponded to the ited much lower diversity than the one obtained from Col Actinobacteriaclass: onewithhighhomologytoMicrobac- duMidi. IntheColduDomesampleonlythreerepresenta- terium thalassium (Richert et al., 2007) and the other with tive sequences were retrieved. Two corresponded to uncul- Pseudonocardia sp. (Fig. 6) which was isolated from ma- tivated Alphaproteobacteria clones, AP-12 and O15B-H01, rinesediments(NCBIaccessionnumberAY974793,unpub- thefirstdetectedinamarineestuary(NCBIaccessionnum- lished). ber AY145551, unpublished) and the other from a uranium Seven sequences retrieved from the aerosol sample from minegroundwater(NCBIaccessionnumberAY662032,un- Antarctica were identified by phylogenetic analysis. Three published). The other sequence corresponded to the class sequences corresponded to the Alphaproteobacteria class: Actinobacteria, with a high level of homology with the one related with clone B3NR69D12, which has been also speciesDietziakujamensis,whichwasisolatedfromtheHi- identifiedintheAndesandtheAlps(ColduMidi); another malaya(Mayilrajetal.,2006). related with clone AP-12 which has been also identified in www.biogeosciences.net/6/33/2009/ Biogeosciences,6,33–44,2009 38 E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil Fig.3. PhylogenetictreeofAgrobacteriumandrelatedbacteria. Cloned16SrRNAsequencesfromenrichmentculturesareindicatedin bold. SequencesretrievedfromNevadoIllimani(Andes)aredesignatedbytheword“Illimani”followedbythenumberofthesequence. SequencesfromColduMidi(Alps)aredesignatedby“Coldumidi”followedbythenumberofthesequence. Barrepresents10%estimated phylogeneticdivergence. Fig.4. PhylogenetictreeofHydrogenophagaandrelatedbacteria. Cloned16SrRNAsequencesfromenrichmentculturesareindicatedin bold. SequencesretrievedfromNevadoIllimani(Andes)aredesignatedbytheword“Illimani”followedbythenumberofthesequence. SequencesfromColduMidi(Alps)aredesignatedby“Coldumidi”followedbythenumberofthesequence. Barrepresents10%estimated phylogeneticdivergence. Biogeosciences,6,33–44,2009 www.biogeosciences.net/6/33/2009/ E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil 39 Fig.5. PhylogenetictreeofAquiflexusandrelatedbacteria. Cloned16SrRNAsequencesfromenrichmentculturesareindicatedinbold. SequencesretrievedfromNevadoIllimani(Andes)aredesignatedbytheword“Illimani”followedbythenumberofthesequence.Sequences fromColduMidi(Alps)aredesignatedby“Coldumidi”followedbythenumberofthesequence.Barrepresents10%estimatedphylogenetic divergence. Fig.6. PhylogenetictreeofPseudonocardiaandrelatedbacteria. Cloned16SrRNAsequencesfromenrichmentculturesareindicatedin bold. SequencesretrievedfromNevadoIllimani(Andes)aredesignatedbytheword“Illimani”followedbythenumberofthesequence. Sequences from aerosol sample collected in Antarctica are designated by “Aeroplankton” followed by the number of the sequence. Bar represents10%estimatedphylogeneticdivergence. www.biogeosciences.net/6/33/2009/ Biogeosciences,6,33–44,2009 40 E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil Table2.AnalysisofOTUs.Microorganismsandsequencescloselyrelatedwithclonesobtainedinthisstudyandsomeoftheircharacteris- tics. ColduDome(snow– ColduMidi(snow– Illimani(snow– UruguayAntarcticSta- French Antarctic Sta- Interestingcharacteristics NCBIa theAlps) theAlps) theAndes) tionArtigas(soil–King tion Dumont d’Urville GeorgeIsland) (aerosol) CloneAP-12 CloneAP-12 CloneAP-12 Unculturedbacterium AY145551(98%) NearBradyrhizobium NearBradyrhizobium NearBradyrhizobium fromWeserEstuary Alphaproteobacteria Alphaproteobacteria Alphaproteobacteria CloneB3NR69D12 CloneB3NR69D12 Unculturedbacterium AY186080(99%) NearAfipiamassiliensis NearAfipiamassiliensis fromaninhabiting Alphaproteobacteria Alphaproteobacteria ferromanganese depositsinLechuguilla andSpiderCaves CloneO15B-H01 Unculturedbacterium AY662032(100%) NearSinellagranuli fromagroundwater Alphaproteobacteria contaminatedwithhigh levelsofnitric acid-bearinguranium water CloneIrT-J614-14 CloneIrT-J614-14 Unculturedbacterium AJ295675(99%) NearAgrobacterium NearAgrobacterium fromauranium Alphaproteobacteria Alphaproteobacteria miningwastepiles Clone Brevundimonassp. Brevundimonas –Unculturedbacterium DQ336984(97%) EV818CFSSHH29 Alphaproteobacteria vesicularis fromsubsurface DQ177489(99%) NearBrevundimonas Alphaproteobacteria waterofKalahariShield, AY169433(99%) Alphaproteobacteria SouthAfrica –Bacteriaisolatedfrom permafrost. –Anaerobicpsychrophilic enrichmentcultures obtainedfroma Greenlandglaciericecore -Bacteriadetectedin lakesnowaggregatesby FISH CloneD-15 CloneD-15 CloneD-15 Unculturedbacterium AF522999(99%) NearLimnobacter NearLimnobacter NearLimnobacter from Betaproteobacteria Betaproteobacteria Betaproteobacteria naturalmineralwater Hydrogenophaga Hydrogenophaga Bacteriadetectedinlake AF078769(98%) palleronii palleronii snowaggregatesbyFISH Betaproteobacteria Betaproteobacteria aAccessionNumberinNCBIoftheclosestphylogeneticrelativesandpercentageofsimilarity. ColduMidi;andathirdonecorrespondingtotheBrevundi- regions. Saharan dust layers are very common in high alti- monasvesicularis,whichwasisolatedfromaGreenlandice tude alpine snow (De Angelis AND Gaudichet, 1991). The core(Sheridanetal., 2003). Onesequencecorrespondedto blocks of Alps snow used in this study even recorded a Sa- Betaproteobacteria and had a high level of homology with haradustevent. Dustconcentrationwasalsorelativelyhigh theunculturedcloneofLimnobacterD-15,alreadyidentified inthesnowdepositedonthesummitofIllimani(Clapperton, intheenrichmentculturesofsnowsamplesfromtheAndes 1993), despite the absence of bare rocks near the sampling andtheAlps(ColduMidi). Threesequencescorresponded site. Similar results of microbial abundances in snow were to the Actinobacteria class: one related with Pseudonocar- also demonstrated from various Antarctic localities (Wynn- diaantarctica(Fig.3),apsychrophilicmicroorganismsiso- Williams,1991). lated from McMurdo Valley in the Antarctica (Prabahar et The sample from marine Antarctica soil was a pure cul- al., 2004); a second one corresponding to a member of the ture because only one sequence was retrieved with a high Pseudonocardia genus (Fig. 6) isolated from marine sedi- levelof16SrRNAgenesequencehomologywithamember mentsandalsoidentifiedintheAndes;andathirdonesim- oftheBrevundimonassp.identifiedpreviouslyinpermafrost ilartoBrachybacteriumconglomeratum,whichwasisolated samplesandclosetotheAlphaproteobacteriaspeciesofBre- fromaspacecraftassemblyfacility(NCBIaccessionnumber vundimonasidentifiedintheAntarcticaerosol. AY145551,unpublished). All the identified microorganisms in the enrichment cul- ThehighmountainsnowsfromtheAlpsandAndeswere tures belong to the bacterial domain. Snow samples from richerinbacterialdiversitythantheAntarcticaerosol(Elster the Alps (Col du Midi) and the Andes (Illimani), as well etal.,2007).Thisobservationagreeswiththerelativelylarge as the Antarctic aerosol sample exhibited a similar level of amountsofdusttransportedinthefreetroposphereofthese bacterial diversity in the corresponding enrichment cultures Biogeosciences,6,33–44,2009 www.biogeosciences.net/6/33/2009/ E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil 41 Table2.Continued. ColduDome(snow– ColduMidi(snow– Illimani(snow– UruguayAntarcticSta- French Antarctic Sta- Interestingcharacteristics NCBIa theAlps) theAlps) theAndes) tionArtigas(soil–King tion Dumont d’Urville GeorgeIsland) (aerosol) CloneLCP-79 Unculturedbacterium AF286035(98%) Pseudomonas frommarinesediments Gammaproteobacteria Dietziaspp. Bacteriaisolatedincold DQ060378(99%) Actinobacteria environments(Himalaya andarticOcean) Pseudonocardiasp. Pseudonocardia –Bacteriaisolatedfrom AY234532(99%) Actinobacteria antarctica marinesediments AJ576010(99%) Actinobacteriaand (depthsof500m). Pseudonocardiasp. –Bacteriaisolatedfrom Actinobacteria McMurdo Dry Valleys, Antarctica. Filamentous andproducebrowncolour substratemyceliaand aerialmyceliaVich formawhite conglomerate. Microbacterium Inyoungcultures,cells AM181507(98%) thalassium aresmallirregularrods. Actinobacteria Inoldcultures,rods become shorterorspherical elements. Coloniesareyellowish while,yellow ororange. Brachybacteriumsp. Bacteriaisolatedfrom AY167842(99%) Actinobacteria spacecraftassembly facilities Aquiflexumbalticum Aquiflexumbalticum Bacteriumisolatedfrom AJ744861(94%) Bacteroidetes Bacteroidetes surfacewaterofthe Central BalticSea(depthof5m). Redandtransparent colonieswhenyoung,but turn opaque with ongoing in- cubation. Cellscontaincarotenoids. (Table 2). The marine Antarctic soil (King George Island, Antarcticmaritimesoil),Hydrogenophagapalleroni(Coldu UruguayAntarcticStationArtigas)enrichmentculturecon- Midi and Illimani), Aquiflexus balticus (Col du Midi and tainedonlyonesequence(Table2). Thisisprobablydueto Illimani) and Pseudonocardia sp. (Illimani and Antarctic thestrictoligotrophicprotocolusedforitsenrichment. Only aerosol) (Table 2). Six out of fourteen identified genera or onetypeofbacteriawasabletogrowonmineralagarplate. species are found only in a single location (Table 2). In- On the contrary, in the case of snow and aerosol samples, terestingly enough, much more diversity was found in very growthinliquidmineralmediawasalwaysobserved. Inthe close locations in the Alps (Col du Dome and Col du Midi, supplementary information http://www.biogeosciences.net/ only one common sequence out of ten) than in locations 6/33/2009/bg-6-33-2009-supplement.pdf figures of more in different hemispheres, altitudes or matrix (snow versus phylogenetictreeareprovided. aerosol) (Table 2). Concerning the pigments that color to The distribution of detected bacteria was related to cold someofthecolonies,severalbacteriasuchasDietzia,Aqui- environments (21.4%), marine environments (35.7%) and flexum, Microbacterium and Pseudonocardia, related with soils/subsurface (35.7%) (Table 2). No single cosmopoli- thosepresentintheenrichmentcultures,havebeendescribed tan bacterium was found in all the analyzed sites, although as having this peculiar phenotypic property. In addition, three geographically distant sampling sites (Alps, Illimani membersoftheActinobacteriaclassarewellknownfortheir and Antarctic aerosol) shared three microorganisms (Lim- ability to synthesize pigments for radiation protection pur- nobacter clone D-15, and two uncultured Alphaproteobac- poses. teria, clones B3NR69D12 and AP-12). Different bacteria It is evident that the experimental procedure used in this werefoundintwolocations:Agrobacteriumsp.(ColduMidi study is not contamination-free (see Elster et al., 2007). and Illimani), Brevundimonas sp. (Antarctic aerosol and Careful analysis of contamination was done through the www.biogeosciences.net/6/33/2009/ Biogeosciences,6,33–44,2009 42 E.Gonza´lez-Toriletal.: Diversityofpigmentedbacteriainaerosol,snowandsoil use of blank samples (e.g. vials without sample but merely References opened in the field for the same period of time, distilled or re-distilled water used throughout the project, filters for Achenbach, L. and Woese, C.: 16S and 23S rRNA-like primers, inArchaea, in: Alaboratorymanual, editedby: Sower, R.and aerosolcollection,negativecontrolsforDNAextractionand Schreier,H.J.,ColdSpringHarborLaboratoryPress,NY,521– PCR amplification, etc.). No growth could be detected in 523,1995. anyofthecontrolsperformed. Inaddition,mostoftheiden- Bischoff,H.W.andBold,H.C.:SomealgaefromEnchantedRock tified bacteria fit quite well with bacteria isolated or identi- andrelatedalgalspecies,inPhycologicalstudies,Universityof fiedfromsimilarhabitatsaroundtheworld(e.g.Castroetal., Texas,IV,Austin,68,1963. 2004; Shivaji et al., 2004; Mayilraj et al., 2006; Despre´s et Brettar, I., Christen R., and Ho¨fle M. G.: Aquiflexum balticum al.,2007;Georgakopoulosetal.,2008).Thesebacterialcom- gen.nov.,sp.nov.,anovelmarinebacteriumoftheCytophaga- munitiesrepresentindividualsfrequentlyoccurringinremote Flavobacterium-Bacteroides group isolated from surface water terrestrial cold or hot deserts/semi-deserts, and/or marginal of the central Baltic Sea, Int. J. Syst. Evol. Microbiol., 54(6), soil-snow-iceecosystems. 2335–2341,2004. Concerningthedifferentmodelsforlongdistancemicro- Brosius,J.,Dull,T.J,Sleeter,D.D.,andNoller,H.F.: Geneorga- nizationandprimarystructureofaribosomalRNAoperonfrom bial dissemination (atmospheric circulation, ocean currents, Escherichiacoli,J.Mol.Biol.,148,107–127,1981. birds,fish,mammalsandhumanvectors),ifweconsiderthe Castro,V.A.,Thrasher,A.N.,Healy,M.,Ott,C.M.,andPierson, habitatsinwhichrelatedmicroorganismshavebeendetected, D.L.: Microbialcharacterizationduringtheearlyhabitationof we can rule out ocean currents and animal vectors, leaving theinternationalspacestation,MicrobialEcology,47,119–126, atmospheric circulation as the most plausible means of dis- 2004. semination,especiallygiventhemicrobialdiversitydetected Clapperton, C. C.: Quaternary Geology and Geomorphology of in the Antarctic aerosol and shared with other distant loca- South America, in Elsevier Science Publ., Amsterdam, 780, tions(71.4%),whichunderlinestheimportanceofmicrobes 1993. attachedtodustparticlesfortheirdisseminations. Chalmers, M. O., Harper, M. A., and Marshall, W. A.: An Il- Of course, our data can not rule out the possible model lustrated Catalogue of Airborne Microbiota from the Maritime of“everythingiseverywhere”,althoughconsideringtheex- Antarctic,BritishAntarcticSurvey,Cambridge,175,1996. Christner, B. C., Mosley-Thompson, E., Thompson, L. G., perimental constraints introduced by the use of extremely Zagorodnoc, V., Sandman, K., andReeve, J.N.: Recoveryand oligotrophicselectivemedia,wethinkthatatmosphericdis- identification of viable bacteria immured in glacial ice, Icarus, persioncanexplainadequatelytheobservedresults. Oneof 144,479–485,2000. theproblemsrelatedwiththistypeofexperimentsistorule De Angelis, M. and Gaudichet, A.: Saharan dust deposition over out the possibility of contamination. Considering the loca- MontBlanc(FrenchAlps)duringthelast30years,Tellus,43B, tion of the selected sampling sites and the obtained results, 61–75,1991. we believe we demonstrated by the protocols used that this Despre´s,V.R.,Nowoisky,J.F.,Klose,M.,Conrad,R.,Andreae,M. possibilityisimprobable. Althoughthedifferencesobserved O.,andPo¨schl,U.:Characterizationofprimarybiogenicaerosol among the samples obtained during the same campaign in particles in urban, rural, and high-alpine air by DNA sequence twoclosesitesintheAlpsisnotobvious,wethinkthatitis andrestrictionfragmentanalysisofribosomalRNAgenes,Bio- a very useful internal control for the lack of contamination geosciences,4,1127–1141,2007, http://www.biogeosciences.net/4/1127/2007/. inthesamplingmanipulationbecausetheywerecollectedby Dimmick,R.L.,Wolochow,M.A.,andChatigny,A.A.: Evidence thesameteamandanalyzedtogetherinthesameconditions. formorethanonedivisionofbacteriawithinairborneparticles, We strongly believe that the common microbial patterns of Appl.Environ.Microbiol.,38,642–643,1979. the particular microorganisms observed at distant locations Du,H.,Jiao,N.,Hu,Y.,andZeng,Y.:Diversityanddistributionof reflectstrueairbornemicrobialdissemination,andtherefore pigmentedheterotrophicbacteriainmarineenvironments,FEMS wewouldliketoproposetheprotocolofenrichmentinolig- MicrobialEcology,57,92–105,2006 otrophicmediaforfurthertestingofairbornemicrobialdis- Elster,J.,Lukesˇova´,A.,Svoboda,J.,Kopecky´,J.,andKanda,H.: persiontoreducetheamountofcontaminantsthatcouldin- Diversityandabundanceofsoilalgaeinthepolardesert,Sver- terferewiththeanalysisoftheresults. drup pass, central Ellesmere Island, Polar Rec. 35, 231–254, 1999. Acknowledgements. The work was supported by grants Elster,J.andBenson,E.E.: LifeinthePolarTerrestrialEnviron- from the Spanish Ministerio de Educacio´n y Ciencia (grant ment: AFocusonAlgae,inLifeInTheFrozenState,FullerB, CGL2006/02534/BOS), the Ministry of Education of the editedby: Lane,N.andBenson,E.E.,TaylorandFrancis,Lon- Czech Republic (Kontakt – ME 934, ME 945 and project GA don,111–149,2004. CR 206/05/0253) and the French-Czech cooperative research Elster,J.,Delmas,R.J.,Petit,J.-R.,andReha´kova´,K.: Composi- programme Barrande No. 99054. We are very grateful to tionofmicrobialcommunitiesinaerosol,snowandicesamples Mrs.JanaSˇnokhousova´forhertechnicalassistance. fromremoteglaciatedareas(Antarctica,Alps,Andes),Biogeo- sciencesDiscuss.,4,1779–1813,2007, Editedby:J.Toporski http://www.biogeosciences-discuss.net/4/1779/2007/. Flechtner, V. R.: Enigmatic desert soil algae. Soil algal flora of Biogeosciences,6,33–44,2009 www.biogeosciences.net/6/33/2009/
Description: