The RNA Sensor RIG-I Dually Functions as an Innate Sensor and Direct Antiviral Factor for ... PDF
Preview The RNA Sensor RIG-I Dually Functions as an Innate Sensor and Direct Antiviral Factor for ...
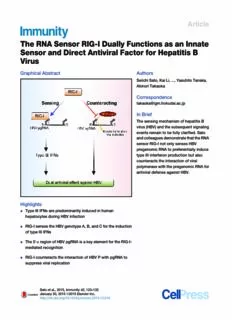
Article The RNA Sensor RIG-I Dually Functions as an Innate Sensor and Direct Antiviral Factor for Hepatitis B Virus Graphical Abstract Authors SeiichiSato,KaiLi,...,YasuhitoTanaka, AkinoriTakaoka Correspondence [email protected] In Brief ThesensingmechanismofhepatitisB virus(HBV)andthesubsequentsignaling eventsremaintobefullyclarified.Sato andcolleaguesdemonstratethattheRNA sensorRIG-InotonlysensesHBV pregenomicRNAtopreferentiallyinduce typeIIIinterferonproductionbutalso counteractstheinteractionofviral polymerasewiththepregenomicRNAfor antiviraldefenseagainstHBV. Highlights d TypeIIIIFNsarepredominantlyinducedinhuman hepatocytesduringHBVinfection d RIG-IsensestheHBVgenotypeA,B,andCfortheinduction oftypeIIIIFNs d The50-εregionofHBVpgRNAisakeyelementfortheRIG-I- mediatedrecognition d RIG-IcounteractstheinteractionofHBVPwithpgRNAto suppressviralreplication Satoetal.,2015,Immunity42,123–132 January20,2015ª2015ElsevierInc. http://dx.doi.org/10.1016/j.immuni.2014.12.016 Immunity Article The RNA Sensor RIG-I Dually Functions as an Innate Sensor and Direct Antiviral Factor for Hepatitis B Virus SeiichiSato,1,2,10KaiLi,1,2,10TakeshiKameyama,1,2,10TakayaHayashi,3,10YujiIshida,4ShukoMurakami,5 TsunamasaWatanabe,5SayukiIijima,5YuSakurai,6KoichiWatashi,7SusumuTsutsumi,5YusukeSato,6HidetakaAkita,6 TakajiWakita,7CharlesM.Rice,8HideyoshiHarashima,6MichinoriKohara,9YasuhitoTanaka,5andAkinoriTakaoka1,2,* 1DivisionofSignalinginCancerandImmunology,InstituteforGeneticMedicine,HokkaidoUniversity,Sapporo,Hokkaido060-0815,Japan 2MolecularMedicalBiochemistryUnit,BiologicalChemistryandEngineeringCourse,GraduateSchoolofChemicalSciencesand Engineering,HokkaidoUniversity,Sapporo,Hokkaido060-0815,Japan 3ResearchCenterforInfection-AssociatedCancer,InstituteforGeneticMedicine,HokkaidoUniversity,Sapporo,Hokkaido060-0815,Japan 4PhoenixBioCo.,Ltd.,Higashihiroshima,Hiroshima739-0046,Japan 5DepartmentofVirologyandLiverUnit,NagoyaCityUniversityGraduateSchoolofMedicalSciences,Nagoya,Aichi467-8601,Japan 6LaboratoryofInnovativeNanomedicine,FacultyofPharmaceuticalSciences,HokkaidoUniversity,Sapporo,Hokkaido060-0812,Japan 7DepartmentofVirologyII,NationalInstituteofInfectiousDiseases,Tokyo208-0011,Japan 8LaboratoryofVirologyandInfectiousDisease,TheRockefellerUniversity,NewYork,NY10065,USA 9DepartmentofMicrobiologyandCellBiology,TokyoMetropolitanInstituteofMedicalScience,Setagaya-ku,Tokyo156-8506,Japan 10Co-firstauthor *Correspondence:[email protected] http://dx.doi.org/10.1016/j.immuni.2014.12.016 SUMMARY zer et al., 2012; Revill and Yuan, 2013). Around four hundred million people worldwide are persistently infected with HBV, Host innate recognition triggers key immune re- whichisamajorcausativefactorassociatedwithnotonlyinflam- sponses for viral elimination. The sensing mecha- mationbutalsocirrhosisandevencanceroftheliver.Currently, nism of hepatitis B virus (HBV), a DNA virus, and interferon(IFN)andnucleoside/nucleotideanalogsareavailable thesubsequentdownstreamsignalingeventsremain for HBV treatment (Rehermann and Nascimbeni, 2005; Hale- to be fully clarified. Here we found that type III but goua-De Marzio and Hann, 2014). However, the long-term responseratesarestillnotsatisfactory.Elucidationofhostim- not type I interferons are predominantly induced in muneresponseagainstHBVinfectioniscrucialforbetterunder- human primary hepatocytes in response to HBV standing of the pathological processes and viralelimination to infection, through retinoic acid-inducible gene-I controlHBVinfection. (RIG-I)-mediated sensing of the 50-ε region of HBV ThetypeIIFNs,IFN-aandIFN-b,arerepresentativecytokines pregenomicRNA.Inaddition,RIG-Icouldalsocoun- thatelicithostinnateimmuneresponsesagainstviralinfections. teract the interaction of HBV polymerase (P protein) Inaddition,anotherIFNfamily,typeIIIIFNs(IFN-l,alsoknownas with the 50-ε region in an RNA-binding dependent IL-28andIL-29)exhibitspotentantiviralactivitysimilartoIFN-a manner,whichconsistentlysuppressedviralreplica- andIFN-b(Sheppardetal.,2003;Kotenko,2011;Kotenkoetal., tion.Liposome-mediateddelivery andvector-based 2003).ProductionoftypeIandtypeIIIIFNsismassivelyinduced expression of this ε region-derived RNA in liver inmanytypesofcellsuponinfectionwithvariousviruses,which abolished the HBV replication in human hepato- isknowntobemediatedbytheactivationofpattern-recognition receptors (PRRs). During virus infection, virus-derived nucleic cyte-chimeric mice. These findings identify an acids(bothRNAandDNA)aremainlysensedbycertainPRRs, innate-recognitionmechanismbywhichRIG-Idually suchasretinoicacid-induciblegene-I(RIG-I)(Yoneyamaetal., functions as an HBV sensor activating innate 2004;Choietal.,2009;Chiuetal.,2009;Ablasseretal.,2009), signaling and to counteract viral polymerase in melanomadifferentiation-associatedgene5(MDA5)(Yoneyama humanhepatocytes. etal.,2005),cyclicGMP-AMPsynthase(cGAS)(Sunetal.,2013), andIFN-g-inducibleprotein16(IFI16)(Unterholzneretal.,2010). Particularly, RIG-I is a key PRR that can detect virus-derived INTRODUCTION RNAsinthecytoplasmduringinfectionwithavarietyofviruses, such as influenza virus, hepatitis C virus (HCV), and measles HepatitisBvirus(HBV)isahepatotropicvirusoftheHepadnavir- virus,whicharecloselyrelatedtohumandiseasepathogenesis idae family and contains a circular, partially double-stranded (RehwinkelandReiseSousa,2010).BindingofRIG-Itoitsligand DNA genome of about 3.2 k base pairs that is replicated via RNAs, such as 50-triphosphorylated RNA or short double- reverse transcription of a pregenomic RNA (pgRNA). HBV stranded RNAs (Takeuchi and Akira, 2009; Hornung et al., causes hepatic inflammation associated with substantial 2006), activates the downstream signaling pathways in a morbidityworldwide(RehermannandNascimbeni,2005;Prot- mannerdependentontheadaptorproteinmitochondrialantiviral Immunity42,123–132,January20,2015ª2015ElsevierInc. 123 signalingprotein(MAVS;alsoknownasIPS-1,VISA,orCardif) cytes(PHH)invitro24hrafterinfectionwithHBV-C(Figure1C); (TakeuchiandAkira,2009),leadingtotheinductionoftheIFN- however,neitheroftypeInortypeIIIFNtestedwasinduced(Fig- regulatoryfactor-3(IRF-3)andNF-kB-dependentgeneexpres- uresS1DandS1E).Althoughitisdifficulttosimplycomparethe sionandthesubsequentproductionoftypeIandtypeIIIIFNs amountofIFNinducedbydifferenttypesofvirus,theinductionof and inflammatory cytokines (Takeuchi and Akira, 2009). Thus, IFN-l1, l2, and l3 mRNAs in response to HBV infection was RIG-I sensing of viral RNA is a crucial process to activate the muchweakerthanthatofNewcastlediseasevirus(NDV)infec- antiviralinnateresponsestolimitviralreplicationandtheactiva- tion(Figure1C).Inthisregard,inordertoruleoutthepossibility tionofadaptiveimmunity(TakeuchiandAkira,2009). thattheIFN-lresponseisduetocontaminantsintheinocula,we Asforthevirusesthatareknowntobetheleadingcauseofhe- usedLamivudine (LAM),anHBV inhibitor, in thisassay. Treat- paticinflammation, RIG-IisthemajorPRRthatinitiates innate mentwithLAMinhibitedIFN-lmRNAinductioninresponseto immuneresponsesagainstHCV.RIG-IsensingofHCVismedi- HBV infection in PHH (Figure 1C), suggesting that the IFN atedthroughitsrecognitionofthepoly-U/UCmotifoftheHCV response is actually induced by HBV replication. Furthermore, RNAgenome30nontranslatedregion,whichleadstotheactiva- weanalyzedHepG2-sodium taurocholatecotransporting poly- tionoftypeIIFNresponse(Saitoetal.,2008).Ontheotherhand, peptide (NTCP)-C4 cell line (Iwamoto et al., 2014) stably ex- earlierstudieshaveshownthattheinnateimmuneactivationis pressing human NTCP, a functional receptor for HBV (Yan impaired and the induction of type I IFNs such as IFN-a or etal.,2012),andconfirmedthatIFN-l1andIFN-induciblegenes IFN-bishardlydetectedinanimalmodelsofHBVinfection,as suchasOAS2andRSAD2,butnotIFN-b,wereinducedinthese comparedwithHCVinfection(Wielandetal.,2004;Nakagawa cellsafterinfectionwithHBV-C,andthattheseinductionswere et al., 2013). However, it is still not fully clarified how HBV is abolishedbytreatmentwithLAM(Figure1D).Tonextassessthe recognizedbyhumanhepatocytesandtheroleoftypeIIIIFNs innateimmuneresponsesinvivoduringHBVinfection,weex- aswell. ploitedseverecombinedimmunodeficiencymicethatcarrythe HerewereportthatHBVinfectionpredominatelyinducestype urokinase-type plasminogen activator transgene controlled by III,butnottypeI,IFNgeneinduction,whichismediatedbyRIG-I an albumin promoter (uPA+/+/SCID mice), in which more than throughitsrecognitionofthe50-εregionofHBV-derivedpgRNA. 70% of murine hepatocytes were replaced by human hepato- WealsoshowthatRIG-IcancounteracttheinteractionofHBV cytes (Tateno et al., 2004) (hereinafter referred to as chimeric polymerase (P protein) with the 50-ε region of pgRNA in an mice).Afterthechimericmicewereintravenouslyinfectedwith RNA-binding dependent manner, resulting in the suppression HBV-C,whichwasderivedfrompatientswithchronichepatitis, of HBV replication. Furthermore, liposome-mediated delivery theexpressionoftypeIIIIFNmRNAsincreasedinthelivertissue, and expression of the 50-ε region-derived RNA in liver sup- whereas IFN-a4and IFN-bmRNAs were not upregulated (Fig- pressedtheHBVreplicationinvivoinchimericmicewithhuman- ure 1E). In parallel with this type III IFN response, we also ized livers. Thus, our findings demonstrate the innate defense observed the expression of IFN-inducible genes, such as mechanismsbasedontheviralRNA-RIG-Iinteraction,whereby CXCL10,OAS2,andRSAD2,inthehumanliveroftheseinfected RIG-IfunctionsnotonlyasaHBVsensorfortheactivationofIFN mice(Figure1E).ThesefindingsindicatethatamoderatetypeIII responsebutalsoasadirectantiviralfactor. butnottypeIortypeIIIFNresponseisactivatedinhumanhepa- tocytesinresponsetoHBVinfection. RESULTS HBV-InducedTypeIIIIFNExpressionDependsonRIG-I TypeIIIIFNsArePredominantlyInducedinHepatocytes Wenextdeterminedwhichsensor-mediatedsignalingpathway duringHBVInfection is responsible for the HBV-induced type III IFN response. As To investigate the innate immune activation during HBV infec- HBVisaDNAvirus(RehermannandNascimbeni,2005;Protzer tion,weexaminedtypeIandtypeIIIIFNresponsesinhumanhe- etal.,2012;RevillandYuan,2013),weassessedthecontribution patocytes.Consistentwiththepreviousreports(Wielandetal., of previously reported cytosolic DNA sensors including RIG-I 2004;Nakagawaetal.,2013),wehardlyobservedtheinduction (Chiuetal.,2009;Ablasseretal.,2009;Choietal.,2009),IFI16 oftypeIIFNs,IFN-a4,andIFN-binresponsetotransfectionwith (Unterholzneretal.,2010),andcGAS(Sunetal.,2013)inhuman plasmids carrying 1.24-fold the HBV genome of three major hepatocytes.KnockdownanalysesrevealedthatIFN-l1induc- different genotypes, Ae (HBV-Ae), Bj (HBV-Bj), and C (HBV-C) tion in HepG2 or Huh-7 cells by plasmid transfection for (Figure1AandFigureS1Aavailableonline)atleastuptoseven HBV-CorHBV-Ae,respectively,wassuppressedbytheknock- days after transfection, although the expression of HBV RNAs downofRIG-I,butnotthatoftheothersensors(Figures2A,S2A was detectable (Figure S1B). On the other hand, type III IFN, andS2B).To furtherconfirm theinvolvement of RIG-Iin HBV- IFN-l1,wasinducedinallofthethreetypesofhumanhepato- triggered type III IFN response, we measured IFN-l1 mRNA cyte cell line tested (Figures 1A and S1A). In HepG2 cells, expression induced by plasmid expression in Huh-7.5 cells HBV-C shows the highest IFN-l1 response, which was also thatcarryadominant-negativemutantRIG-Iallelethatprevents confirmed byELISA, albeit weakly (Figures 1Aand 1B). More- RIG-I signaling (Saito et al., 2007), as compared with Huh-7 over,IFN-l1inculturesupernatantcouldinhibitvesicularstoma- cells that have an intact RIG-I pathway. Huh-7.5 cells failed titisvirus(VSV)replicationinplaquereductionassay,aswellas to induce IFN-l1 mRNA expression in response to HBV-Ae HBV replication (Figure S1C), indicating the physiological rele- genomeplasmidtransfection,asinthecaseofstimulationwith vance of the induced IFN-l1 to antiviral activities. Consistent 50-triphosphate RNA (3pRNA), a RIG-I ligand (Takeuchi and withtheseresults,weobservedthesignificantinductionofnot Akira,2009; Hornungetal.,2006) (Figure2B).Inconcordance onlyIFN-l1butalsoIFN-l2and-l3inprimaryhumanhepato- withthisresult,knockdownoftripartitemotifcontainingprotein 124 Immunity42,123–132,January20,2015ª2015ElsevierInc. Figure1. IFN-lInductioninHumanHepatocytesinResponsetoHBVInfection (A)QuantitativeRT-PCR(qRT-PCR)analysisofIFNL1(left),IFNA4(middle),andIFNB1(right)mRNAattheindicatedtimesaftertransfectionwith1.24-foldthe HBVgenome(genotypeAe,Bj,orC)oremptyvector(Mock)inHepG2cells. (B)ELISAofIFN-l1at48or72hraftertransfectionwiththeHBVgenomeinHepG2cells.Thedotlineindicatestheminimumdetectableamount(31.2pg/ml)of IFN-l1bytheELISAkit.ND,notdetected,indicatesbelowtheminimumdetectableamount. (C)qRT-PCRanalysisofIFNL1,IFNL2,andIFNL3mRNAat24hrafterinfectionwithHBV,NDV(multiplicityofinfection=10)ormock((cid:2)),ormedia-treated Lamivudine(LAM)ascontrolinprimaryhumanhepatocytes(PHH).ThemRNAcopynumber(±SD)ofeachsubtypeoftypeIIIIFNper1mgtotalRNAuponHBV-C infectionisasfollows:IFNL1(83,197.6±6,241.4)andIFNL2/3(409,280.6±119,676.2). (D)TimecourseanalysesbyqRT-PCRofIFNL1,OAS2,RSAD2,IFNB1mRNA,andpgRNAattheindicatedtimesafterHBVinfectioninHepG2-hNTCP-C4cells. TheeffectofLamivudinetreatmentwasalsoanalyzed. (E)qRT-PCRanalysisofIFNL1,IFNL2,IFNL3,IFNA4,IFNB1,CXCL10,OAS2,andRSAD2mRNAoflivertissuesat4or5weeksafterinfectionwithHBV-Cin chimericmice.((cid:2)),uninfectedmice.Redlinesrepresentthemeanofeachdataset.*p<0.05and**p<0.01versuscontrol.RE,relativeexpression.(A–D)Dataare presentedasmeanandSD(n=3)andarerepresentativeofatleastthreeindependentexperiments.SeealsoFigureS1. 25(TRIM25),MAVS,TANK-bindingkinase1(TBK1),andIRF-3, sultedinthesuppressionofIFN-l1mRNAinductioninHepG2 all of which are signaling molecules essentially involved in the cells in response to transfection with the HBV-C genome. On RIG-I-mediated IFN pathway (Takeuchi and Akira, 2009), re- theotherhand,suchaneffectwasnotobservedincellstreated Immunity42,123–132,January20,2015ª2015ElsevierInc. 125 Figure2. RIG-I-DependentIFN-lInduction inResponsetoHBVInfection (A)HepG2cellstreatedwithcontrolsiRNA(Con- trol)orsiRNAtargetingRIG-I,IFI16,orcGASwere transfected with the HBV-C genome for 48 or 72hr.TheamountofIFN-l1weremeasuredby ELISA.Thedotlineindicatestheminimumcyto- kineexpressiondetected(31.2pg/ml)ofIFN-l1by theELISAkit.ND,notdetected,indicatesbelow detectableconcentrations(left),andknockdown efficiency was analyzed by immunoblotting (IB) (right). (B)qRT-PCRanalysisofIFNL1mRNAinHuh-7or Huh-7.5 cells transfected with the HBV-Ae genome(at24hraftertransfection)orstimulated with3pRNA(1mg/ml)for6hr. (C)HepG2cellstreatedwithcontrolsiRNA(Con- trol)ortheindicatedsiRNAsweretransfectedwith the HBV-C genome. At 48 hr after transfection, totalRNAsweresubjectedtoqRT-PCRanalysis forIFNL1. (D)qRT-PCRanalysisofIFNL1mRNAinsiRNA- treatedPHHat24hrpostinfectionwithindicated HBVgenotype.Mock,emptyvector-transfected. ((cid:2)), uninfected. Data were normalized to the expression of GAPDH. Data are presented as meanandSD(n=3)andarerepresentativeofat least three independent experiments. *p < 0.05 and **p < 0.01 versus control in (B) or HBV-in- fectedcontrolgroupin(A,C,andD).NS,notsig- nificant.SeealsoFigureS2. witheitherTRIF(alsoknownasTICAM-1)orMYD88siRNA(Fig- the expression of all of these RNA transcripts and tested its ures2CandS2C).Inaddition,weconfirmedthattheknockdown effect on HBV-induced IFN-l1 expression. As shown in Fig- ofRIG-IandMAVSabolishedIFN-l1inductioninPHHinfected ure 3B, knockdown with this siRNA (Figure S3A) suppressed witheachgenotype(Figure2D).Furthermore,wealsoconfirmed IFN-l1inductioninHuh-7cellstransfectedwithHBV-Ae.Next, by knockdown assay that the induction of IFN-l1 and OAS2 todeterminewhichoftheseHBVRNAtranscriptsis/areinvolved mRNAinHepG2-hNTCP-C4cellsinresponsetoinfectionwith intheRIG-I-mediatedIFN-l1induction,wepreparedexpression HBV-CwasdependentonRIG-I(FigureS2E).Thesedataindi- vectors to express each of these four viral transcripts in catethatIFN-l1geneinductionduringHBVinfectiondepends HEK293T cells that are often used to analyze RIG-I signaling largelyonRIG-Isignalingpathway. pathway in human cells. As a result, it is only the longest 3.5kbtranscript,thatis,pgRNA,thathasthepotentialtoelicit The50-εRegionofHBVpgRNAIsaKeyElementfor a significant induction of IFN-l1 mRNA (Figures 3C and S3B). RIG-I-DependentIFN-l1Induction ItwasalsoconfirmedbyknockdownanalysiswithpgRNA-tar- RIG-Icanrecognizenotonlyvirus-derivedRNAbutalsoDNAin geted siRNA, which showed significant suppression of IFN-l1 the cytoplasm (Yoneyama et al., 2004; Choi et al., 2009; Chiu inductioninHepG2cellstransfectedwithHBV-Ae(FigureS3C). etal.,2009;Ablasseretal.,2009).TofurtherclarifyhowRIG-I Theseresultssuggestthat50-1.1kbregionofHBVpgRNAiscrit- recognizes HBV, we first examined either or both of which icalfortheactivationofRIG-IpathwaytoinduceIFN-l1expres- nucleic acid (DNA and RNA) derived from HBV-infected cells sion.Ontheotherhand,theremainingthreetranscripts,which canactivateIFN-l1geneexpression.Transfectionwithnucleic alsocontainthesamesequenceofpartofthis1.1kbregionof acidfractionsextractedfromHBVinfectedHuh-7cellsafterpre- HBV pgRNA at the 30 end of their transcripts, failed to induce treatment with RNase A, but not DNase I resulted in marked IFN-l1mRNA(Figure3C).AnartificiallydeletedformofpgRNA, inhibition of the IFNL1 promoter activation, suggesting that which lacks this overlapping sequence at the 30-region (D3), virus-derivedRNAsmightbecandidatesoftheRIG-Iliganddur- showed IFN-l1 induction, whereas such response was not ingHBVinfection(Figure3A). observedfor anothermutant pgRNAlacking itatthe 50-region The HBV genome comprises a partially double-stranded (D5)(Figure3D).Thesedataalsosupportapossibleimportant 3.2kbDNA.DuringalifecycleofHBVinhepatocytes,itscova- role of the 50-overlapping sequence of HBV pgRNA for RIG-I- lentlyclosedcircularDNA(cccDNA)istranscribedtogenerate mediatedIFN-l1induction. fourmajorRNAspecies:the3.5,2.4,2.1,and0.7kbviralRNA The50-endofHBVpgRNAisknowntocontaintheencapsida- transcripts (Rehermann and Nascimbeni, 2005; Protzer et al., tionsequence,called‘‘epsilon(ε),’’whichtakesastem-loopsec- 2012;RevillandYuan,2013).WecreatedansiRNAtosuppress ondary structure (Junker-Niepmann et al., 1990; Pollack and 126 Immunity42,123–132,January20,2015ª2015ElsevierInc. Figure3. RIG-IActivationIsMediatedbyIts Recognition of the 50-ε Region of HBV pgRNA (A) Luciferase activity of an IFN-l1 reporter plasmid after 24 hr of stimulation with nucleic acids (2 mg/ml) extracted from Huh-7 cells transfected with control plasmid (Mock) ortheHBV-AegenomewithorwithoutRNaseA or DNase I treatment. RLU, relative luciferase units. (B) Huh-7 cells treated with control or HBV RNA-targeted siRNA were transfected with the HBV-Ae genome or mock. After 24 hr of trans- fection,totalRNAsweresubjectedtoqRT-PCR forIFNL1. (CandD)Aschematicrepresentationoffourtypes ofHBVRNAs,pgRNA(3.5kb),2.4kb,2.1kb,and 0.7 kb RNAs in (C), and two deleted forms of pgRNA,D5andD3,in(D).Theoverlappingregion is shown in blue. qRT-PCR analysis of IFNL1 mRNAofHEK293Tcellsafter24hroftransfection withtheindicatedexpressionvectors.Datawere normalized to the amount of each HBV RNA expression(CandD). (E)AschematicrepresentationofpgRNA,εRNA, or control RNA (ContRNA) (left). HEK293T cells treated with control or RIG-I siRNA were unstimulated (Mock) or stimulated with εRNA for 12 hr. Total RNAs were subjected to qRT- PCRforIFNL1(middle).qRT-PCRanalysisofIFNL1mRNAinHEK293Tcellsafter12hrofstimulationwithεRNAorContRNA(right).EachoftheRNAswas preparedbyinvitrotranscription. (F)HEK293Tcellsweretransfectedwitheachplasmidforstem-loopmutantsofpgRNA(5STM,3STM,or5and3STM),andthensubjectedtoqRT-PCRanalysis asdescribedin(C).*p<0.05and**p<0.01versuscontrolin(A,B,andE)orversus3.5Kin(C,D,andF).NS,notsignificant.SeealsoFigureS3. Ganem, 1993; Knaus and Nassal, 1993; Jeong et al., 2000). ContRNA,inHEK293Tcells(Figure4A,top).Similarly,endoge- Therefore,wehypothesizedthatthis50-εstructuremightconfer nousRIG-IinteractedwithεRNAalbeitweakly(Figure4A,bot- apossiblepathogen-associatedmolecularpattern(PAMP)motif tom). We also demonstrated the intracellular colocalization of for RIG-I recognition. To test this hypothesis, we stimulated RIG-IwithεRNAinHuh-7.5cells(Figure4B).Inaddition,RNA- HEK293TandHepG2cellswiththeεregion-derivedRNA(here- binding protein immunoprecipitation (RIP) assay revealed that after called εRNA). Consequently, IFN-l1 mRNA was signifi- thefulllengthofHBVpgRNAwasdetectedintheRIG-I-immuno- cantly induced, which was dependent on RIG-I, while such a precipitatedcomplex,andD5pgRNAandD3pgRNAwerealso responsewasnotdetecteduponstimulationwiththeequivalent detected(FigureS4A),whichisseeminglyinconsistentwiththe length of RNA that is derived from HBV pgRNA but does not results by the functional assay (Figures 3C, 3D, 3F and S3C). containanyεelement(ContRNA)(Figure3EandS3D).Wealso Theseresultssuggestthattheεregionisrequiredforitsinterac- confirmedRIG-I-dependentIRF-3activationinresponsetostim- tionwithRIG-I,butonlythe50-εregionisnecessarytoactivate ulationwithεRNA(FiguresS3DandS3E).Duetotheoverlapping RIG-I pathway. We further tried to determine which region of sequenceof50-and30-endsofHBVpgRNAasmentionedabove, RIG-ImediatesitsinteractionwithHBVpgRNA.BothRIPassay thisεelementisfoundatbothendsofpgRNA.Wenextgener- andRNApull-downassaywithseveraldeletionmutantsofRIG-I atedseveralmutantformsofHBVpgRNA,eachofwhichcarries showedthattheC-terminalportionofRIG-I(C-RIG)includingits mutationswithin50-or30-εregionorbothtodisruptthestem- helicasedomainandrepressordomain(RD)exceptforCARDs loop structure (5STM, 3STM, or 5 and 3STM, respectively). In canbindtoHBVpgRNA(Figure4C;FiguresS4BandS4C).In concordancewiththeresultsshowninFigures3Cand3Dand addition, gel shift assay showed that the interaction of HBV S3B, IFN-l1 mRNA induction was detected upon expression εRNA or pgRNA was impaired with the RD or C-RIG mutant, ofthe3STMtranscriptthathasanintact50-εregion,assimilar respectively, each of which carries a point mutation (K888E) to that of intact 3.5-kb pgRNA (Figure 3F). In contrast, either that abolishes its RNA-binding activity (Cui et al., 2008) (Fig- 5STMor5and3STMdidnotshowsignificantresponse.These ure4D).AsimilarresultwasalsoobtainedbyRIPassay,wherein findings indicate that the 50-ε region of HBV pgRNA is critical thewild-type(WT)C-RIG,butnottheK888Emutant,wascoim- forIFN-l1inductionpossiblythroughtherecognitionbyRIG-I. munoprecipitatedwithHBVpgRNA(FigureS4D),likeHCVRNA thatwaspreviouslyreportedtointeractwithRIG-I(FigureS4E). RIG-IInteractswiththeε-RegionofpgRNA WealsoconfirmedtheinteractionofHBVpgRNAwithendoge- Next,weassessedtheinteractionofRIG-Iwiththeεregionof nousRIG-IinHepG2cells,whereasitsinteractionwithothernu- HBV pgRNA, that is, εRNA. Pull-down assays showed that cleicacidsensors,suchasIFI16andMDA5(Yoneyamaetal., Flag-tagged RIG-I was coprecipitated with εRNA,but notwith 2005), was not detected (Figure 4E). These data indicate that Immunity42,123–132,January20,2015ª2015ElsevierInc. 127 Figure4. RIG-IInteractswiththeεRegion ofpgRNA (A) RNA pull-down assay showing the binding activityoftheindicatedRNAstoFlag-taggedRIG-I (Flag-RIG-I)inHEK293Tcells(top)orendogenous RIG-IinHepG2cells(bottom). (B)FRETanalysisfortheinteractionofYFP-tagged RIG-I (YFP-RIG-I) with rhodamine (ROX)-conju- gatedεRNA(εRNA-ROX)orContRNA(ContRNA- ROX). Representative fluorescence images of YFP,ROX,andFRETC/YFP(theratioofcorrected FRET(FRETC)toYFP).Arrowheadsindicatearea showing high FRET efficiency. Scale bar repre- sents20mm.Right,dotplotofFRETC/YFPratio (smallhorizontalbars,mean). (C)RIPassaywithHEK293Tcelllysatesexpress- ingseveralFlag-taggeddeletionmutantsofRIG-I andpgRNAexpressionvectorbyusinganti-Flag antibody.ImmunoprecipitatedpgRNAwasquan- titatedbyqRT-PCRandnormalizedtotheamount ofimmunoprecipatedproteins(FigureS4C)andis represented as fraction of input RNA prior to immunoprecipitation(percentageinput). (D) Gel-shift analysis of complex formation be- tweenεRNAandrecombinantRIG-IRD(WT)orRD (K888E).Arrowheadsdenotepositionofunbound RNAandRNA–RIG-Icomplexes. (E) RIP assay with HepG2 cell lysates prepared after 48 hr of transfection of the HBV-C genome by using anti-RIG-I, anti-IFI16, anti-MDA5, or control immunoglobulinG.TheimmunoprecipitatedpgRNAwasmeasuredbyqRT-PCR(top)asdescribedin(C).Whole-cellexpressionandimmunoprecipitated amountsofRIG-I,IFI16,andMDA5(bottom).DataarepresentedasmeanandSD(n=3)andarerepresentativeofatleastthreeindependentexperiments. *p<0.05and**p<0.01versuscontrolin(BandE).NS,notsignificant.SeealsoFigureS4. the50-εregionofviralpgRNAfunctionsasanHBV-associated K888E(Cuietal.,2008)mutantcouldnotinhibitthebindingof molecular pattern to be specifically recognized by RIG-I and PproteinwithpgRNA(Figures5DandS5C).Inaddition,treat- cantriggerIFN-lresponse. mentwithrecombinantIFN-l1inHuh-7.5cellsupregulatedthe amountofthemutantRIG-Iprotein(T55I)(FigureS5D),resulting RIG-IExertsanAntiviralActivitybyCounteractingthe inapartialinhibitionofthePproteininteractionwithpgRNA,and InteractionofHBVPolymerasewithpgRNA this inhibitory effect was abrogated by RIG-I knockdown (Fig- WenextassessedthecontributionofRIG-Ipathwayinantiviral ure5E).Infact,FRETanalysisshowedthatthePprotein-εRNA defense against HBV infection. RIG-I knockdown in PHH re- interaction was significantly suppressed by expression of the sulted in a higher HBV genome copy number at 10 days after RIG-IRD(WT)alone,butnotthemutantRD(K888E)(FigureS5E). infection with HBV-C, as compared with PHH treated with Furthermore,HBVreplicationwasalsosuppressedbyexpres- control siRNA (Figure 5A). A similar observation was made for sionoftheRIG-IRD(WT)inHuh-7.5cells,whereinanyIFNinduc- RIG-I siRNA-treated HuS-E/2 cells (Figure S5A). These results tionisnotobserved,whilethemutantRD(K888E)didnotaffect indicateanimplicatedroleofRIG-Iasaninnatesensortoacti- viral replication (Figure 5F). These findings revealed another vate antiviral response against HBV infection. On the other aspectofRIG-Iasadirectantiviralfactorthroughitsinterference hand, it has been previously reported that the 50-ε region of withthebindingofHBVPproteintopgRNAinanIFNpathway- HBVpgRNAisimportanttoserveasabindingsiteofviralPpro- independentmanner. tein for initiating reverse transcription (Bartenschlager and Schaller, 1992). As consistentwith this, weshowed that the P TheεRNARestrictsHBVReplicationinHuman protein interacts with εRNA in Huh-7.5 and HEK293T cells, by Hepatocyte-ChimericMice fluorescence resonance energy transfer (FRET) analysis (Fig- Lastly,basedontheaboveresults,wetriedtoharnessthether- ure 5B) and RNA pull-down assay (Figure S5B), respectively. apeuticpotentialofthePprotein-interactingεRNAforthecontrol These findings facilitated us to examine whether RIG-I could ofHBVinfection.Avectorwasdesignedtoincludea63bpDNA block the access of P protein toward the ε region. As we ex- oligo, which is transcribed into an εRNA. We confirmed in the pected, recombinant RIG-I protein suppressed the interaction invitroexperimentsusingHuh-7.5cellsthatεRNAinducedby of P protein with pgRNA in a dose-dependent manner (Fig- thisvector-drivenexpressioniscapabletofunctionasadecoy ure5C).SuchaninhibitoryeffectwasalsoobservedinHuh-7.5 RNA to interfere with the binding of HBV P protein to pgRNA cells by expression of WT RIG-I, as well as its T55I (Sumpter andtoinhibitviralreplicationinanIFN-independentmanner(Fig- etal.,2005;Saitoetal.,2007)orK270A(Takahasietal.,2008) ures6Aand6B,left).Ontheotherhand,εRNAdidnotshowany mutant (Figures 5D and S5C), both of which are not able to difference in HCV replication as compared with control (Fig- induceligand-dependentactivationofthedownstreamsignaling ure 6B, right). In order to evaluate the therapeutic efficacy of but retain their RNA-binding activities. On the other hand, the εRNA in vivo, we exploited HBV infection model of human 128 Immunity42,123–132,January20,2015ª2015ElsevierInc. Figure 5. RIG-I Functions as an Antiviral FactorbyCounteractingtheInteractionof HBVPProteinwithpgRNA (A)qPCRanalysisofcopynumbersofencapsi- dated HBV DNA (left) and qRT-PCR analysis of RIG-I(middle)andIFNL1mRNA(right)incontrolor RIG-IsiRNA-treatedPHHafter10daysofinfection withHBV-C. (B)FRETanalysisfortheinteractionbetweenYFP- taggedPprotein(YFP-P)orYFPandεRNA-ROX asdescribedinFigure4B.Scalebarrepresents 20 mm. Arrowheads indicate area showing high FRETefficiency. (C)HEK293TcelllysatesexpressingpgRNAand HA-taggedPprotein(HA-P)wereincubatedwith the indicated amount of recombinant RIG-I (rRIG-I).TheinteractionofpgRNAwithHA-Pwas analyzedbyRIPassayandqRT-PCRanalysisas describedinFigure4C. (D)CelllysatesfromHuh-7.5cellsexpressingHBV pgRNA, HA-P,andFlag-RIG-Ioritsmutantsas indicated were subjected to RIP assay for the characterization of the capability of RIG-I to counteracttheinteractionofpgRNAwithHA-P,as describedinFigure4C. (E)TheeffectofrIFN-l1treatmentontheinterac- tion of pgRNA with HA-P in Huh-7.5 cells was assessedbyRIPassay.Huh-7.5cellsexpressing bothpgRNAandHA-PweretreatedwithrIFN-l1 (100ng/ml)for24hr,andsubjectedtoRIPassay asdescribedinFigure4C.RIG-Idependencywas alsodeterminedbyRIG-Iknockdownanalysis. (F) Huh-7.5 cells were transfected with an expressionvectorforRIG-IRD(WT)orRD(K888E), togetherwiththeHBV-Aegenome.After72hrof transfection,copynumbersofencapsidatedHBV DNA were measured (left), as described in (A). ExpressionofFlag-RIG-IRD(WT)andRD(K888E) (right).*p<0.05and**p<0.01versuscontrol.NS, notsignificant.SeealsoFigureS5. hepatocyte-chimericmice.HBV-infectedmiceunderwentintra- loopofHBVpgRNA(Figures1,2,3,and4).Inthisrespect,there venousadministrationwiththeεRNAexpressionvectorloaded havealsobeenseveralreportsshowingthatHBVXorPprotein inaliposomalcarrier,amultifunctionalenvelope-typenanode- interactswithMAVSorcompetesforDDX3bindingwithTBK1, vice (MEND) for efficient delivery, for 2 weeks. Treatment with respectively (Wei et al., 2010; Wang and Ryu, 2010; Yu et al., εRNA-MENDsignificantlysuppressedtheelevationofthenum- 2010), and inhibits RIG-I-mediated type I IFN pathway, which berofviralgenomecopiesintheserabylessthanonetenthof possibly enables HBV to evade from antiviral innate immune thoseforcontrolmice(Figure6C).Consistently,immunofluores- response.ThiswouldmirrortheimportantroleofRIG-I-mediated cenceanalysesshowedthattheexpressionofHBVcoreantigen signaling for antiviral defense against HBV infection, although (HBcAg) in the liver tissues of εRNA-MEND-treated chimeric furtherinvestigationwillberequiredtodeterminewhetherother micewasremarkablyreducedascomparedwiththoseofcontrol sensingmoleculesexceptforRIG-Iareengagedintheactivation mice(Figure6D). of innate responses in other cell types including dendritic cell subsets. Interestingly, Lu et al. have recently showed that the DISCUSSION genotypeDofHBVissensedbyMDA5,butnotRIG-I,whichis basedonlyupontheanalyseswithHBVgenome(2-fold)plasmid Theinnateimmunesystemactsasafrontlineofhostdefense transfectioninasinglecelllineHuh-7(LuandLiao,2013).Inthis against viral infection. In this step, PRRs play a crucial role in respect,wepresumethatsuchseeminglycontradictoryresults the recognition of invading viruses. In particular, nucleic acid mightarisemainlyfromthedifferenceinHBVgenotype:Ithas sensingofvirusesiscentraltotheinitiationofantiviralimmune beenreportedthatthegenotypeDisphylogeneticallydifferent responses.Inthisstudy,wetriedtoseekforarelevantnucleic fromthegenotypesA,B,andC,whichweanalyzedinthisstudy acidsensor(s)forHBVandtocharacterizetheIFNresponsedur- (Katoetal.,2002). ing HBV infection. As a result, we have identified RIG-I as an Inaddition,accordingtoourresults(Figure1CandS1C),HBV- important innate sensor of HBV to predominantly induce type inducedtypeIIIIFNresponsedoesnotseemtobeefficientas IIIIFNsinhepatocytesthroughitsrecognitionofthe50-εstem- comparedwiththecasewithNDVinfection.Wespeculatethat Immunity42,123–132,January20,2015ª2015ElsevierInc. 129 Figure 6. Inhibition of HBV Replication by εRNA (A)Huh-7.5cellsweretransfectedwithexpression vectors for HA-P and εRNA, together with the HBV-Ae genome. RIP assay was performed to evaluate the effect of εRNA on the interaction between HA-P and pgRNA, as described in Figure5D. (B)CopynumbersofencapsidatedHBVDNAin Huh-7.5cellsexpressingtheHBV-Aegenomeand εRNA,asdeterminedbyqPCR(left).HCVrepli- cationinHuh-7.5.1/Rep-Feo-1bcellsexpressing εRNA,asdeterminedbyluciferaseassay(right). (C)HBV-infectedmicewereintravenouslyadmin- istratedwiththeεRNAexpressionvector(εRNA- MEND)oremptyvector(Control-MEND)loadedin liposomalcarrieratadoseof0.5mg/kgofbody weightevery2daysfor14days.SerumHBVDNA inHBV-infectedchimericmicewasdeterminedby qPCR(n=3pergroup).Day0indicatesthetimeof theinitiationofadministration. (D)Immunofluorescenceimagingwasperformed forthedetectionofHBcAg (red)andhumanal- bumin(green)intheliversectionsofHBV-infected chimeric mice at 14 days after treatment with εRNA-MEND or Control-MEND as described in ExperimentalProcedures.Dataarepresentedas meanandSD(n=3)andarerepresentativeofat leastthreeindependentexperiments.**p<0.01 versuscontrol.NS,notsignificant. the weakness of the IFN response during HBV infection might poly-U/UCtractinthe30 nontranslatedregionofHCVgenome attribute at least in part to these viral evasions from host-cell (Saito et al., 2008) and 50 terminal region of influenza virus control, which would be supported by our preliminary data genome (Baum et al., 2010) were previously reported to be showing that one HBV mutant, which generates viral RNAs directly or indirectly critical for viral replication (You and Rice, including pgRNA but lacks the ability to express whole viral 2008;Huang etal.,2005;Moeller etal.,2012). Inthisrespect, proteins including HBV X and P proteins, can induce higher one could envisage that such an exquisite targeting by RIG-I amounts of IFN-l1 than intact HBV (Figure S1G). In relevance would confer a unique machinery to ensure efficient antiviral withthis,ourpresentdataindicatethattheinteraction ofHBV activitiesofRIG-I.Therefore,RIG-Iislikelytoplaydualrolesas P protein with the 50-ε stem-loop affects the RIG-I-mediated aninnatesensorandasadirectantiviraleffectorforhostdefense recognition of viral pgRNA and the subsequent downstream duringviralinfection. signaling events, which might likely suppress the induction of InrelationtotheevaluationoftheexperimentsshowninFig- IFN-ls.ThismightprovideanaspectofHBVPproteininterms ures6Cand6D,weadditionallyanalyzedthefollowingpoints: ofviralevasionfromRIG-Iactivation.Asforthemechanismfor When we treated HepG2 cells with εRNA-MEND or Control- the preferential induction of type III IFNs in hepatocytes in MEND,inbothcaseswehardlydetectedthemassiveinduction responsetoHBV,aswellasHCV(Nakagawaetal.,2013;Park of cytokines such as TNF, IL6, and CXCL10 (data not shown). et al., 2012), we might speculate the existence of a hepato- This was further confirmed by analyzing SCID mice injected cyte-specific factor(s), which is selectively involved in type III withεRNA-MENDorControl-MEND (datanotshown).Inaddi- IFNgeneinduction,althoughthisissuemeritsfurtherinvestiga- tion, εRNA-MEND has the specific effect on the replication of tionincludingepigeneticevaluationofhumanhepatocytes.We HBV, but not HCV in Huh-7.5 cells (Figure 6B). These data also found that either of the 50-or 30-ε region of pgRNA could suggest that the results (Figures 6C and 6D) might not be interactwithRIG-Ibutitwasonlythe50-εregionthatcontributed mainlyinfluencedbymassiveproductionofantiviralcytokines, totheinductionofIFN-l1(Figures3DandS4A).Inthisrespect, although the cross-reactivity of cytokines should be still care- wepresumethatsomecofactor(s)mightadditionallydetermine fully considered. Therefore, it is presumed that the effect of thepreferentialuseofthe50-εregionforRIG-Iactivation;how- εRNA might be based on not only its antagonistic activity but ever,itwouldbeanextinterestingissuetobesolved.Inaddition also its cytokine-inducing activity. These findings might afford tothis,ourdatademonstratedahitherto-unidentifiedfunctionof a new therapeutic modality in replace of conventional antiviral RIG-IasadirectantiviralfactoragainstHBVinfection(Figure5). drugsthathavebeenreportedtohavearisktodevelopdrug- Mechanistically,RIG-Iwasfoundtocounteracttheaccessibility resistance HBV (Song et al., 2012). The present study might of HBV P protein to the 50-ε stem-loop of pgRNA, which is an provide a better approach to the strategy for development of important process for the initiation of viral replication (Bar- nucleicacidmedicineandofferanattractiveclinicaloptionfor tenschlagerandSchaller,1992).Asisthecasewiththis,several thetherapy againstnotonlyHBVbutalsopossibly other virus viralPAMPsknowntoberecognizedbyRIG-I,forexample,the infections. 130 Immunity42,123–132,January20,2015ª2015ElsevierInc. EXPERIMENTALPROCEDURES AUTHORCONTRIBUTIONS InfectionofHumanHepatocyte-ChimericMicewithHBVandInVivo S.S.,K.L.,T.K.,andT.H.carriedoutmostoftheexperimentsandanalyzed TreatmentwithεRNA data.Y.T.providedmaterialsofHBVanddesignedtheprotocolforinfection. To generate human hepatocyte-chimeric mice, uPA+/+/SCID mice, were Y.I.conductedHBVinfectioninchimericmiceandpreparationofhumanhepa- transplantedwithcommerciallyavailablecryopreservedhumanhepatocytes tocytes,andS.M.,T.Watanabe,S.I.,S.T.,andY.T.performedtherelatedanal- (a 2-year-old Hispanic female; BD Bioscience) as described previously ysisandcontributedtotheinterpretationoftheresults.C.M.R.offeredcritical (Tateno et al., 2004). Chimeric mice were intravenously infected with adviceonthewholemanuscriptandprovidedapairofHuh-7andHuh-7.5cell HBV-C (106copiespermouse)derived frompatient withchronic hepatitis lines.K.W.andT.WakitaprovidedHepG2-hNTCP-C4cells.T.K.carriedout (Sugiyamaetal.,2006).TotalRNAswereisolatedfromlivertissuesat0,4, FRET analysis. Y. Sakurai, Y. Sato, H.A., and H.H. syntheses of plasmid- or 5 weeks after infection and subjected to quantitative RT-PCR (qRT- loaded MEND,andM.K.contributedtoestablishmentofthe protocols for PCR). In preparation for εRNA treatment, HBV infection at 3 weeks after in vivo treatment. A.T. supervised the project, designed experiments and infection was confirmed bymeasuring thenumberofviralgenome copies wrotethemanuscriptwithcriticalinputfromthecoauthors,andallauthors in the sera of HBV-infected chimeric mice by qPCR analysis, and at contributedtodiscussingtheresults. 4-weekpostinfection,εRNAexpressionvectororemptyvectorloadedina liposomalcarrier,amultifunctionalenvelope-typenanodevice(MEND),was ACKNOWLEDGMENTS administered intravenously at a dose of 0.5 mg/kg of body weight (n = 3 pergroup)everytwodaysfor14days.Serumsamplesweresubjectedto WethankM.HijikataandK.ShimotohnoforHuS-E/2cellline,N.Sakamotofor qPCR for the quantification of DNA copy numbers of HBV as described Huh-7.5.1/Rep-Feo-1bcells,J.MiyazakiforpCAGGSvector,A.Miyawakifor previously (Nakagawa et al., 2013). All animal protocols described in this pCAGGS-YFPvector,T.Fujitafortheluciferasereporterplasmidp-55C1BLuc, studywereperformedinaccordancewiththeGuidefortheCareandUse A.TakadaforVSV,H.KidaforNDV,H.IshizuandT.Hirosefortechnicaladvice ofLaboratoryAnimalsand approved bythe Animal Welfare Committee of onRIPassay,andA.Bergthaler,A.Shlomai,T.Saito,andM.Yasudaforcritical PhoenixBioCo.,Ltd. readingofthemanuscriptandhelpfuladvice.Theauthorsaregratefulforfinan- cialsupportsfromtheMinistryofHealth,LabourandWelfareofJapan(Grant- HBVInfectioninHumanHepatocytesandQuantificationof in-AidtoA.T.andY.T.),theMinistryofEducation,Culture,Sports,Scienceand EncapsidatedHBVDNA TechnologyofJapan(Grant-in-AidforScientificResearch[A][25253030]to HBVinfectioninPHHorHepG2-hNTCP-C4cellswasperformedat10or100 A.T., Grant-in-Aid for Scientific Research on Innovative Areas [25115502, genomeequivalentspercell,respectively,inthepresenceof4%PEG8000at 23112701]toA.T.,Grant-in-AidforYoungScientists[B][25870015]toS.S.), 37(cid:3)Cfor24hraspreviouslydescribed(Sugiyamaetal.,2006;Watashietal., IRYOHOJINSHADANJIKOKAI(H.TanakaandN.Takayanagi)toA.T.,the 2013;Iwamotoetal.,2014).Lamivudine(50mM;Sigma)wasaddedinHuh-7 KatoMemorialBioscienceFoundationtoA.T.,theYasudaMedicalFoundation culturemediaduringHBVproduction.HBVDNAwaspurifiedfromintracel- toA.T.,theTakedaScienceFoundationtoA.T.,andTheWaksmanFoundation lular core particles as described previously (Turelli et al., 2004; Fujiwara ofJapantoA.T.Y.I.isanemployeeofPhoenixBio. etal.,2005).Briefly,cellsweresuspendedinlysisbuffercontaining50mM Tris-HCl (pH 7.4), 1 mM EDTA, and 1% NP-40. Nuclei were pelleted by Received:June23,2014 centrifugation at 4(cid:3)C and 15,000 rpm for 5 min. The supernatant was Accepted:December3,2014 adjustedto6mMMgOAcandtreatedwithDNaseI(200mg/ml)andofRNase Published:December31,2014 A (100 mg/ml) for 3 hr at 37(cid:3)C. The reaction was stopped by addition of 10mMEDTAandthemixturewasincubatedfor15minat65(cid:3)C.Aftertreat- REFERENCES mentwithproteinaseK(200mg/ml),1%SDS,and100mMNaClfor2hrat 37(cid:3)C,viralnucleicacidswereisolatedbyphenol:chloroform:isoamylalcohol Ablasser, A.,Bauernfeind,F.,Hartmann,G.,Latz,E.,Fitzgerald,K.A., and (25:24:1) extraction and ethanol precipitation with 20 mg glycogen. Copy Hornung,V.(2009).RIG-I-dependentsensingofpoly(dA:dT)throughthein- numbersofHBVDNAweremeasuredbyqPCRwiththeindicatedprimers duction of an RNA polymerase III-transcribed RNA intermediate. Nat. (TableS3). Immunol.10,1065–1072. qRT-PCRAnalysis Bartenschlager,R.,andSchaller,H.(1992).Hepadnaviralassemblyisinitiated bypolymerasebindingtotheencapsidationsignalintheviralRNAgenome. TotalRNAswereisolatedfromculturecellsorfrozenlivertissueusingIsogen EMBOJ.11,3413–3420. (NipponGene)andweretreatedwithDNaseI(Invitrogen).cDNAwasprepared fromtotalRNAsusingReverTraAce(TOYOBO).qRT-PCRwasperformedus- Baum,A.,Sachidanandam,R.,andGarcı´a-Sastre,A.(2010).Preferenceof ingSYBRPremixExTaq(TAKARA)andanalyzedonaStepOnePlusreal-time RIG-IforshortviralRNAmoleculesininfectedcellsrevealedbynext-genera- PCR system (Applied Biosystems). Detailed information about the primers tionsequencing.Proc.Natl.Acad.Sci.USA107,16303–16308. usedhereisshowninTableS3.Datawerenormalizedtotheexpressionof Chiu,Y.H.,Macmillan,J.B.,andChen,Z.J.(2009).RNApolymeraseIIIdetects GAPDHorHBVRNAsforeachsample. cytosolicDNAandinducestypeIinterferonsthroughtheRIG-Ipathway.Cell 138,576–591. RIPAssay Choi,M.K.,Wang,Z.,Ban,T.,Yanai,H.,Lu,Y.,Koshiba,R.,Nakaima,Y., After 2 hr incubation with the antibody as indicated for immunopre- Hangai,S.,Savitsky,D.,Nakasato,M.,etal.(2009).Aselectivecontribution cipitation, cell lysates were mixed with Protein-G Dynabeads (Invitrogen) oftheRIG-I-likereceptorpathwaytotypeIinterferonresponsesactivated and further incubated for 1 hr with gentle shaking. After washing three bycytosolicDNA.Proc.Natl.Acad.Sci.USA106,17870–17875. times,theprecipitatedRNAswereanalyzedbyqRT-PCRwithappropriate Cui,S.,Eisena¨cher,K.,Kirchhofer,A.,Brzo´zka,K.,Lammens,A.,Lammens, primers to detect the target RNA. The amount of immunoprecipitated K., Fujita, T., Conzelmann, K.K., Krug, A., and Hopfner, K.P. (2008). The RNAsisrepresentedasthepercentileoftheamountofinputRNA(percent- C-terminal regulatory domain is the RNA 50-triphosphate sensor of RIG-I. age input). The detail was described in the Supplemental Experimental Mol.Cell29,169–179. Procedures. Fujiwara,K.,Tanaka,Y.,Paulon,E.,Orito,E.,Sugiyama,M.,Ito,K.,Ueda,R., SUPPLEMENTALINFORMATION Mizokami,M.,andNaoumov,N.V.(2005).NoveltypeofhepatitisBvirusmuta- tion:replacementmutationinvolvingahepatocytenuclearfactor1bindingsite SupplementalInformationincludesfivefigures,fourtables,andSupplemental tandemrepeatinchronichepatitisBvirusgenotypeE.J.Virol.79,14404–14410. ExperimentalProceduresandcanbefoundwiththisarticleonlineathttp://dx. Halegoua-DeMarzio,D.,andHann,H.W.(2014).Thenandnow:theprogressin doi.org/10.1016/j.immuni.2014.12.016. hepatitisBtreatmentoverthepast20years.WorldJ.Gastroenterol.20,401–413. Immunity42,123–132,January20,2015ª2015ElsevierInc. 131
Description: